Dear experts,
I am trying to implement systematics in my model. I am doing it following the example in Include Shape systematics. My code is slightly different since I am using unbinned datasets so I use continuous pdfs instead of Histograms as in the example.
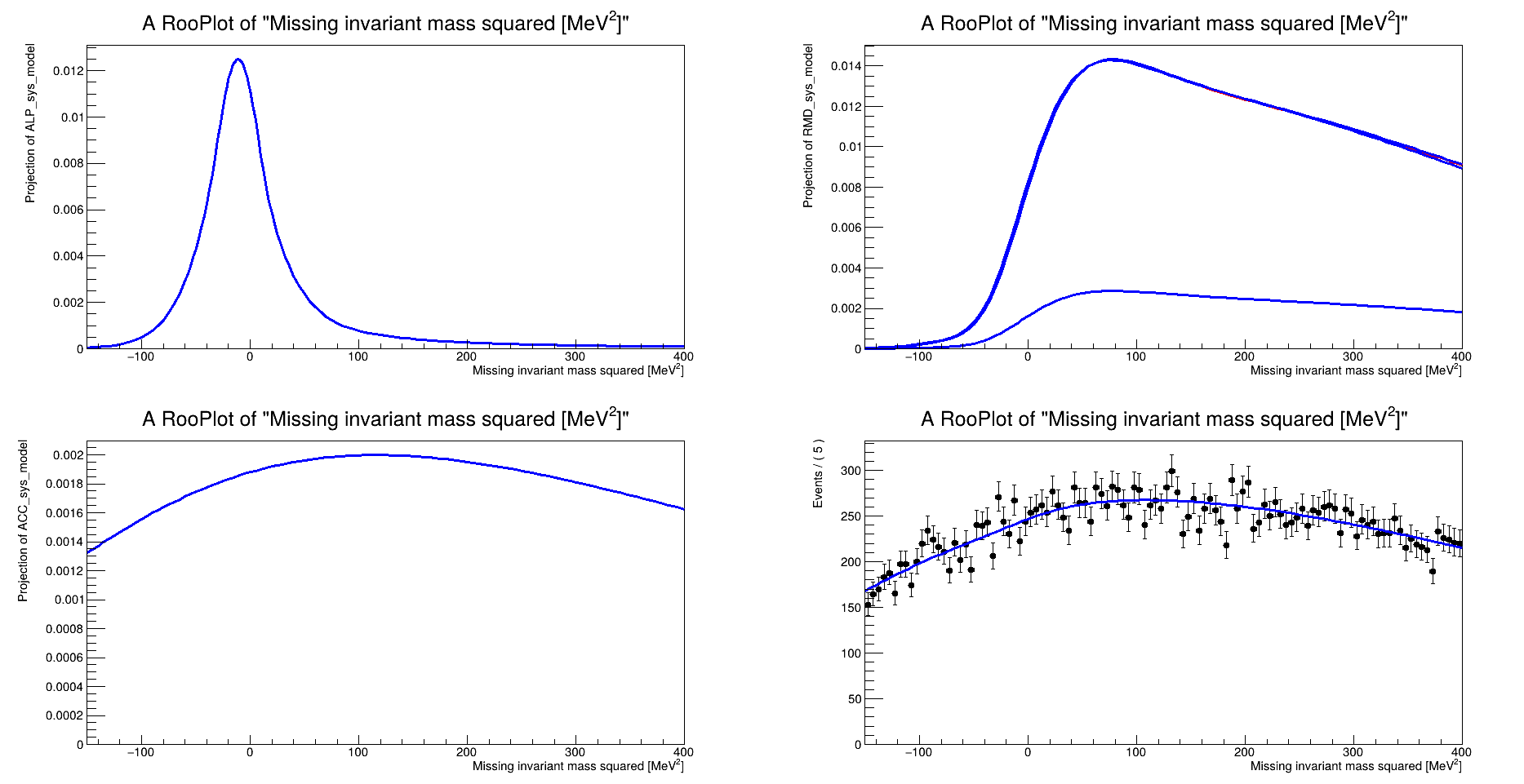
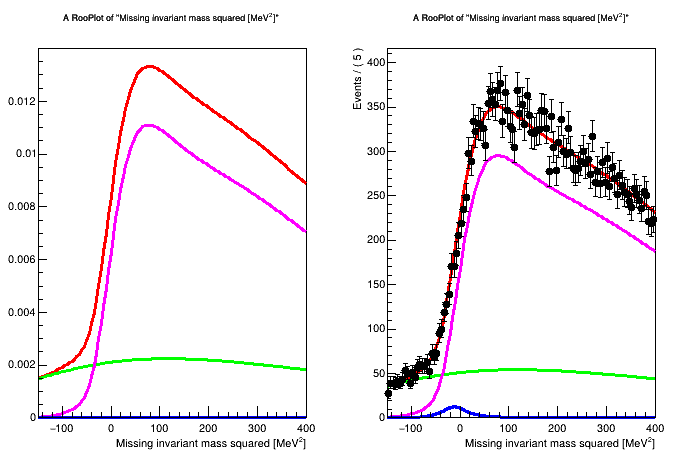
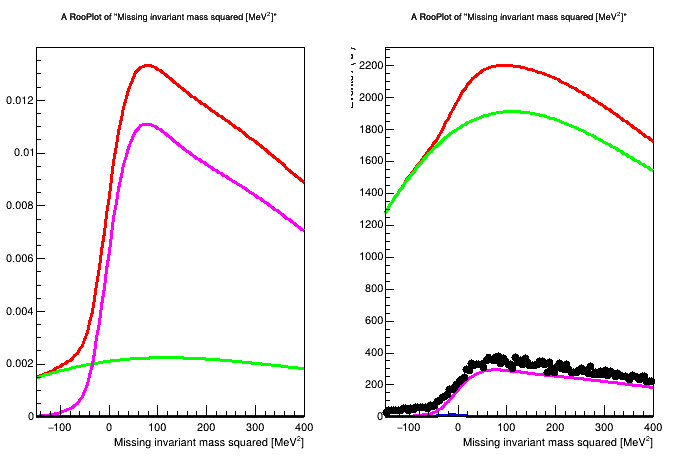
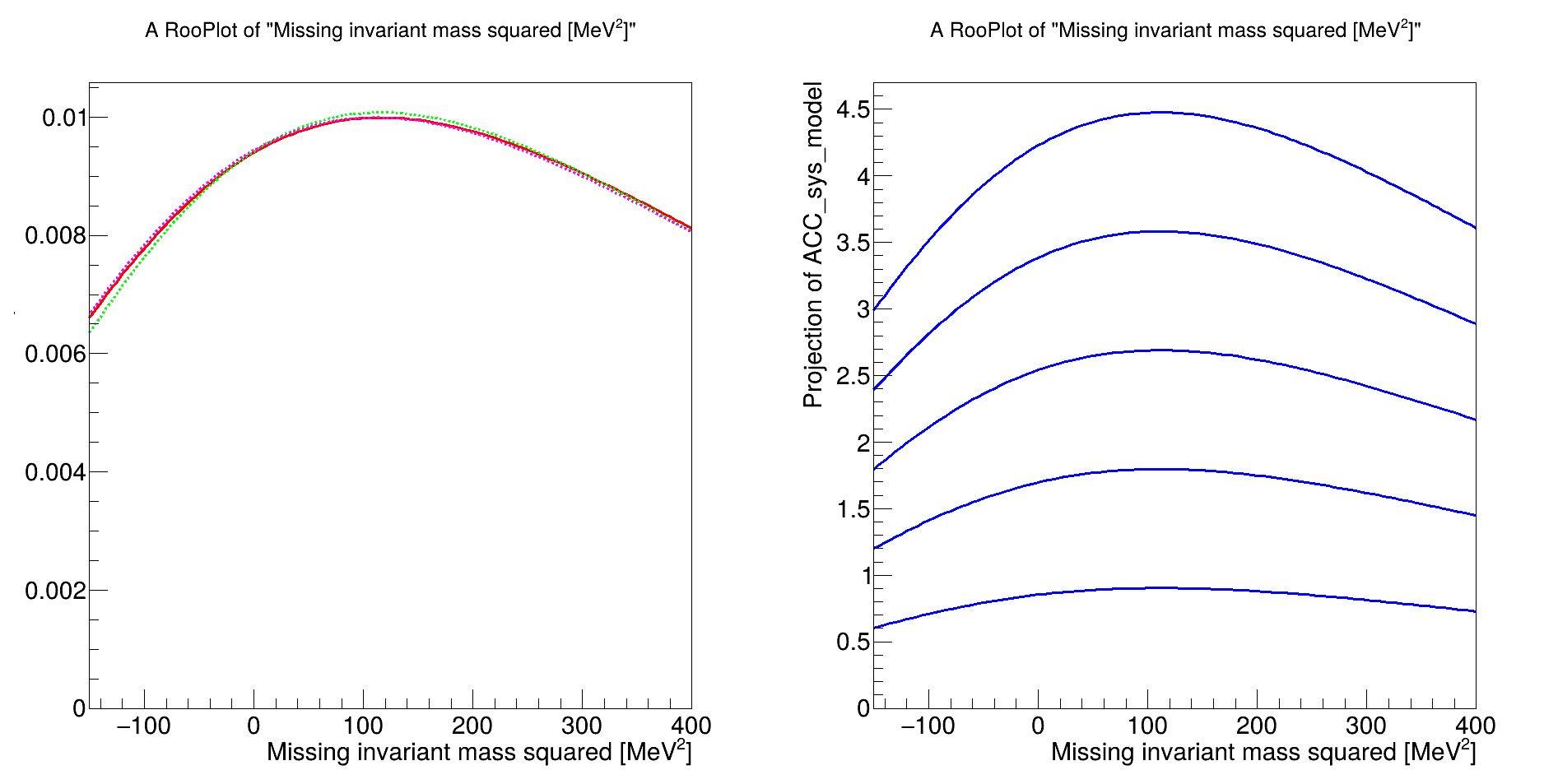
Anyway I define my model as described in the example above (from 3 RooAddPdf and a RooRealVar i construct a PiecewiseInterpolation function) and I get this warning:
WARNING:Eval -- Evaluating RooAddPdf "model_name" without a defined normalization set. This can lead to ambiguous coefficients definition and incorrect results. Use RooAddPdf::fixCoefNormalization(nset) to provide a normalization set for defining uniquely RooAddPdf coefficients!
so I fix my normalization set using my observable in x axis for all my RooAddPdf and I get an error that terminates my code
[#0] ERROR:InputArguments -- RooArgSet::checkForDup: ERROR argument with name "model name"_conditional is already in this set
terminate called after throwing an instance of 'std::runtime_error'
what(): RooAbsCollection::addOwned could not add the argument to the collection! The ownership would not be well defined if we ignore this.
The warning and the error is not triggered if I use RooRealSumPdf instead of RooAddPdf, but in this case the shapes of my pdfs changes (thing that I can manually fix doing something like this Difference between RoorealSum and RooAddPdf but it is a quite nasty given my model)
So I was wondering if it is possible to do with RooAddPdf adjusting something or I must use RooRealSumPdf.
Thanks,
Elia
in the file attached is part of the code where I create my model
question.C (13.9 KB)