Dear all,
I am trying to create a model like this (f*signal + (1-f)*background). Other than the parameter of interest, the signal fraction, I need to include also a nuisance parameter, accounting for the systematics on the background shape.
The background shape variation is defined by a nominal distribution, and the two distributions corresponding to ±1 sigma variations, respectivelly. So I am using the PiecewiaeInterpolation class to model the shape systematic through a parameter alpha.
From this model, I want to generate data, which then I want to use to make fit studies.
The problem I have is that when I set a value of alpha different than zero to generate the data, I get weird behaviour: data and model do not match. What am I missing?
Here is a sample code:
void test_morph()
{
// signal
TH1F* hsgn = new TH1F("hsgn","signal", 20, 0, 100);
TF1* g = new TF1("g","[0]*exp(-(x-[1])^2/(2*[2]^2))", 0, 100);
g -> SetParameters(100, 50, 10);
hsgn -> FillRandom("g", 10000);
// background
TH1F* hbkg_nom = new TH1F("hbkg_nom","nominal bkg", 20, 0, 100);
TF1* fe = new TF1("fe","[0]*exp(-[1]*x)", 0, 100);
fe -> SetParameters(50, 0.02);
hbkg_nom -> FillRandom("fe", 10000);
TH1F* hbkg_p = new TH1F("hbkg_p","+1 sigma bkg", 20, 0, 100);;
fe -> SetParameters(55, 0.03);
hbkg_p -> FillRandom("fe", 10000);
TH1F* hbkg_m = new TH1F("hbkg_m","-1 sigma bkg", 20, 0, 100);;
fe -> SetParameters(45, 0.01);
hbkg_m -> FillRandom("fe", 10000);
// define observables
RooRealVar e{"e", "energy", 0, 100, "MeV"};
e.setBins(Int_t(20));
// signal pdf
RooDataHist dh_sgn{"dh_sgn", "dh_sgn", RooArgSet{e}, hsgn};
RooHistPdf sgn("sgn", "sgn", RooArgSet{e}, dh_sgn);
// background pdf
//
// nominal value
RooDataHist dh_bkg_nom{"dh_bkg_nom", "dh_bkg_nom", RooArgSet{e}, hbkg_nom};
RooHistPdf bkg_0("bkg_0", "bkg_0", RooArgSet{e}, dh_bkg_nom);
//
// bkg +1 sigma
RooDataHist dh_bkg_p{"dh_bkg_p", "dh_bkg_p", RooArgSet{e}, hbkg_p};
RooHistPdf bkg_p("bkg_p", "bkg_p", RooArgSet{e}, dh_bkg_p);
//
// bkg -1 sigma
RooDataHist dh_bkg_m{"dh_bkg_m", "dh_bkg_m", RooArgSet{e}, hbkg_m};
RooHistPdf bkg_m("bkg_m", "bkg_m", RooArgSet{e}, dh_bkg_m);
//
// build interpolation between -1 -> +1 sigma
RooRealVar alpha{"alpha", "alpha bkg sys", 0, -5, 5};
PiecewiseInterpolation bkg_sys("bkg_sys", "bkg_sys", bkg_0, bkg_m, bkg_p, alpha);
// build model
RooRealVar f{"f", "f", 0.5, 0, 1};
RooAddPdf model{"model", "model", RooArgSet{sgn, bkg_sys}, f};
alpha.setVal(0.0);
RooDataHist* data = model.generateBinned(RooArgSet{e}, 1000, RooFit::Verbose(1));
TCanvas* c = new TCanvas();
c->cd();
auto frame = e.frame();
data->plotOn(frame);
model.plotOn(frame, RooFit::LineColor(6));
model.plotOn(frame, RooFit::LineColor(4), RooFit::Components("bkg_sys"));
frame -> Draw();
}
These is what I get with alpha = 0:
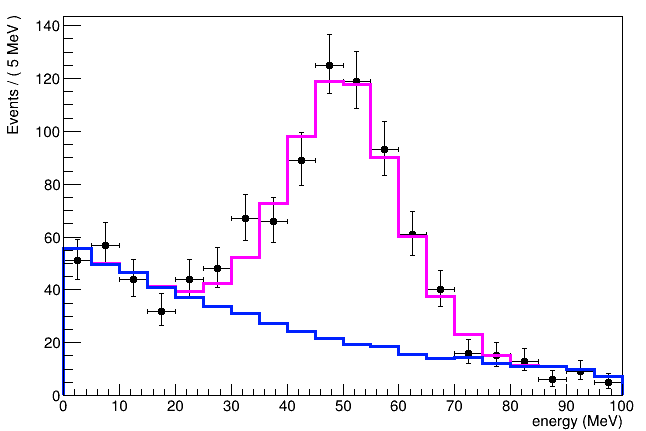
and this the output with alpha > 0:
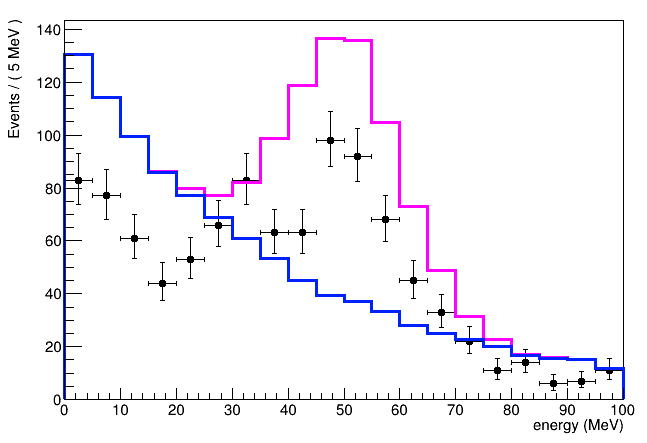
Thanks for your help.
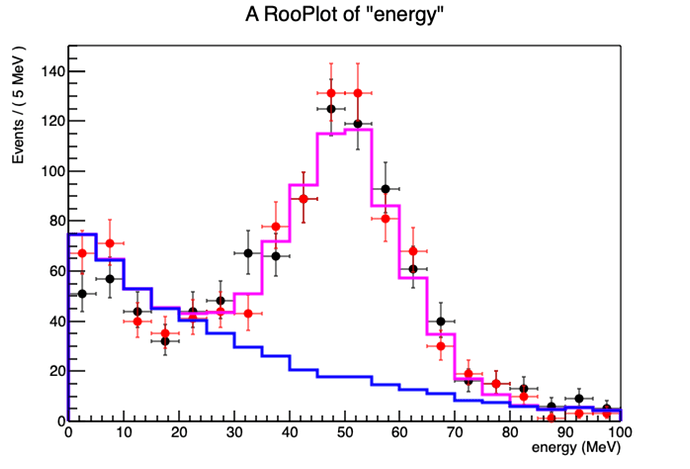
 ). Check if you have components whose sum is below zero.
). Check if you have components whose sum is below zero.