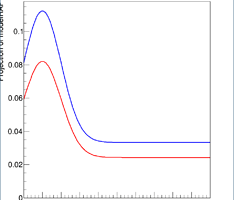
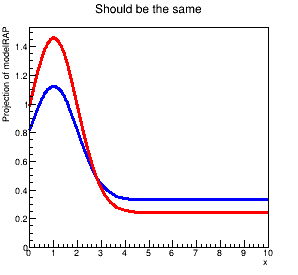
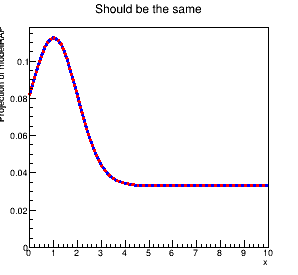
Hi, I’m trying to use an extended RooRealSumPdf, but I get a discrepancy from an extended RooAddPdf. The model is uniform+gaussian. The implementations are:
modelRSP = RooRealSumPdf("modelRSP","modelRSP",
RooArgList(w.pdf("bkg"), w.pdf("sig")),
RooArgList(w.var("bkgNorm"),w.var("sigNorm")),
True
)
And
modelRAP = RooAddPdf("modelRAP","modelRAP",
RooArgList(w.pdf("bkg"), w.pdf("sig")),
RooArgList(w.var("bkgNorm"),w.var("sigNorm")),
)
When I plot these, I get two different results. In particular, even with the same sigNorm value, the RooRealSumPdf seems to have less signal than the RooAddPdf. The plot shows the discrepancy:
I have a stand-alone example of this: errorDemoAddPdfVsSumPdf.py (4.3 KB). Am I missing something here? I’ve tried extending the uniform and gaussian components manually, but I get a similar result. Any help would be very appreciated.