Hi Lorenzo,
I am quite advanced in my project of processing a 2d interpolation using RooMomentMorphND in root. Unfortunately the documentation of this is very poor (or maybe it’s me who could’t find it), so I really start from scratch. So what I am doing is as following:
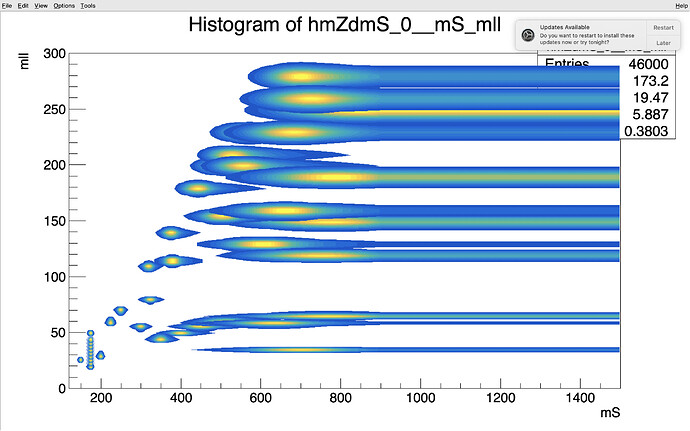
I have 33 h1 histograms and 33 h2 histograms which are 1D histograms. I fit them with a double sided crystall function and save the parameters fits in text file. So my code is reading the text file and make pdfs from the fit parameters. The observable in h1 is mll and the one in h2 is mS. I use PROD:: to construct the pairs (mll, mS) in a 2d plan, wich will give me 33 pairs of (mll, mS). So now I want to populate the 2D plane by interpolating using RooMomentMorphND. The results I got so far are promising as you can in see the file I attached:
The interpolated points are the ones plotted with the option cont1 which form the vertical line along the Y axis at mS ~ 175 GeV.
Here is my code:
#include "RooWorkspace.h"
#include <RooRealVar.h>
#include "RooDataHist.h"
#include "RooHistPdf.h"
#include "RooAbsArg.h"
#include "RooMomentMorphND.h"
#include "RooFit.h"
#include "TRandom3.h"
#include "RooRealVar.h"
#include "RooDataSet.h"
#include "RooGaussian.h"
#include "RooConstVar.h"
#include "RooPolynomial.h"
#include "RooIntegralMorph.h"
#include "RooNLLVar.h"
#include "TCanvas.h"
#include "TAxis.h"
#include "RooPlot.h"
#include "TH1.h"
#include "RooMyPdf.cxx"
using namespace RooFit;
void morphingBackup()
{
TString file_input[]={"./FitParameterM4l_2e2m.txt","./FitParameterAvgMll_2e2m.txt"};
TString tmps;
ifstream fin_mS(file_input[0]);
ifstream fin_mZd(file_input[1]);
int Npoints = 33;
double Param_mS[7][Npoints];
double Param_mZd[7][Npoints];
double PmS[33];
double PmZd[33];
fin_mS>>tmps;fin_mS>>tmps;fin_mS>>tmps;fin_mS>>tmps;fin_mS>>tmps;fin_mS>>tmps;fin_mS>>tmps;fin_mS>>tmps;fin_mS>>tmps;//read first line.
for (Int_t i=0; i<Npoints; i++)
{
fin_mS>>tmps; PmS[i]=tmps.IsFloat()?tmps.Atof():0;
fin_mS>>tmps; PmZd[i]=tmps.IsFloat()?tmps.Atof():0;
fin_mS>>tmps; Param_mS[0][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mS>>tmps; Param_mS[1][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mS>>tmps; Param_mS[2][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mS>>tmps; Param_mS[3][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mS>>tmps; Param_mS[4][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mS>>tmps; Param_mS[5][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mS>>tmps; Param_mS[6][i]=tmps.IsFloat()?tmps.Atof():0;
}
fin_mZd>>tmps;fin_mZd>>tmps;fin_mZd>>tmps;fin_mZd>>tmps;fin_mZd>>tmps;fin_mZd>>tmps;fin_mZd>>tmps;fin_mZd>>tmps;fin_mZd>>tmps;//read first line.
for (Int_t i=0; i<Npoints; i++)
{
fin_mZd>>tmps; PmS[i]=tmps.IsFloat()?tmps.Atof():0;
fin_mZd>>tmps; PmZd[i]=tmps.IsFloat()?tmps.Atof():0;
fin_mZd>>tmps; Param_mZd[0][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mZd>>tmps; Param_mZd[1][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mZd>>tmps; Param_mZd[2][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mZd>>tmps; Param_mZd[3][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mZd>>tmps; Param_mZd[4][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mZd>>tmps; Param_mZd[5][i]=tmps.IsFloat()?tmps.Atof():0;
fin_mZd>>tmps; Param_mZd[6][i]=tmps.IsFloat()?tmps.Atof():0;
}
/* for (int i =0; i< 33; i++)
{
cout << "mS = " << Param_mZd[6][i] << endl;
}*/
RooRealVar mll("mll","mll",0,300) ;
RooRealVar mS("mS","mS", 120, 900) ;
RooArgList pdfs;
TVectorD paramVec = TVectorD(33);
RooWorkspace w("w", 2);
RooAbsPdf *pdf;
TH1 *hmZdmS[33];
TH1 *hmZdmS_morph[10];
//RooBinning dim1(1, 0.0, 1.0);
//RooBinning dim2(1, 0.0, 1.0);
//RooMomentMorphND::Grid referenceGrid(dim1, dim2);
//referenceGrid.addPdf(*gauss1, 0, 0);
for( int i=0; i<33; ++i)
{
w.factory(TString::Format("RooMyPdf::myPdf_mZd_%d(mll[0,300], %f, %f,%f, %f,%f,%f, %f)",i, Param_mZd[0][i],Param_mZd[1][i],Param_mZd[2][i],Param_mZd[3][i],Param_mZd[4][i],Param_mZd[5][i],Param_mZd[6][i]));
w.factory(TString::Format("RooMyPdf::myPdf_mS_%d(mS[120,900], %f, %f,%f, %f,%f,%f, %f)",i, Param_mS[0][i],Param_mS[1][i],Param_mS[2][i],Param_mS[3][i],Param_mS[4][i],Param_mS[5][i],Param_mS[6][i]));
w.factory(TString::Format("PROD::myPdf_mZdmS_%d(myPdf_mZd_%d,myPdf_mS_%d)", i,i,i));
pdf = w.pdf(TString::Format("myPdf_mZdmS_%d",i));
pdfs.add(*pdf);
//referenceGrid.addPdf(*pdf, 0, 0);
paramVec[i]=i;
w.Print() ;
hmZdmS[i] = pdf->createHistogram(TString::Format("hmZdmS_%d",i),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
}
pdfs.Print();
RooPlot* mZdFrame = w.var("mll")->frame();
RooPlot* mSFrame = w.var("mS")->frame();
RooRealVar alpha("alpha","alpha",0,1);
RooArgList varlist;
varlist.add(mS);
varlist.add(mll);
RooArgList parlist;
RooRealVar alpha1("alpha1","alpha1",0,1);
//alpha1=*(w.var("alphaL"));
//parlist.add(*w.var("alphaL"));
TFile f("outputHist.root","recreate");
alpha.setVal(0.1);
RooMomentMorphND morphpdfND_0("morphpdfND_0","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
hmZdmS_morph[0] = morphpdfND_0.createHistogram(TString::Format("hmZdmS_morph_alpha%d",0),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
hmZdmS_morph[0]->Write();
alpha.setVal(0.2);
RooMomentMorphND morphpdfND_1("morphpdfND_1","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
hmZdmS_morph[1] = morphpdfND_1.createHistogram(TString::Format("hmZdmS_morph_alpha%d",0),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
hmZdmS_morph[1]->Write();
alpha.setVal(0.3);
RooMomentMorphND morphpdfND_2("morphpdfND_2","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
hmZdmS_morph[2] = morphpdfND_2.createHistogram(TString::Format("hmZdmS_morph_alpha%d",0),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
hmZdmS_morph[2]->Write();
alpha.setVal(0.4);
RooMomentMorphND morphpdfND_3("morphpdfND_3","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
hmZdmS_morph[3] = morphpdfND_3.createHistogram(TString::Format("hmZdmS_morph_alpha%d",0),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
hmZdmS_morph[3]->Write();
alpha.setVal(0.5);
RooMomentMorphND morphpdfND_4("morphpdfND_4","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
hmZdmS_morph[4] = morphpdfND_4.createHistogram(TString::Format("hmZdmS_morph_alpha%d",0),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
hmZdmS_morph[4]->Write();
alpha.setVal(0.6);
RooMomentMorphND morphpdfND_5("morphpdfND_5","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
hmZdmS_morph[5] = morphpdfND_5.createHistogram(TString::Format("hmZdmS_morph_alpha%d",0),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
hmZdmS_morph[5]->Write();
alpha.setVal(0.7);
RooMomentMorphND morphpdfND_6("morphpdfND_6","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
hmZdmS_morph[6] = morphpdfND_6.createHistogram(TString::Format("hmZdmS_morph_alpha%d",0),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
hmZdmS_morph[6]->Write();
alpha.setVal(0.8);
RooMomentMorphND morphpdfND_7("morphpdfND_7","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
hmZdmS_morph[7] = morphpdfND_7.createHistogram(TString::Format("hmZdmS_morph_alpha%d",0),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
hmZdmS_morph[7]->Write();
alpha.setVal(0.9);
RooMomentMorphND morphpdfND_8("morphpdfND_8","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
hmZdmS_morph[8] = morphpdfND_8.createHistogram(TString::Format("hmZdmS_morph_alpha%d",0),mS, Binning(460,120,1500), YVar(mll,Binning(100)));
hmZdmS_morph[8]->Write();
alpha.setVal(0.9);
hmZdmS[0]->Draw("cont");
for(int i =1; i<33; i++)
{
hmZdmS[i]->Draw("cont same");
hmZdmS[i]->Write();
}
for(int i =0; i<8; i++)
{
hmZdmS_morph[i]->Draw("cont1 same");
}
//hmZdmS_morph[1]->Draw("cont1 same");
f.Close();
}
I varied alpha from 0.1 to 0.9 hoping it will covers the plane but it’s not the case. It’s like alpha is varying in 1 direction along the Y axis. So my question is how should I change alpha to covers the plane? I think I should define a referenceGrid but I don’t know how should I change this line
RooMomentMorphND morphpdfND_8("morphpdfND_8","2D morph",alpha ,varlist, pdfs, paramVec,RooMomentMorphND::Linear);
To include a referenceGrid. Could you advise please?
Tell me if you want to run my code I can give you the other files that are included.
Best regards,
Diallo.