ROOT Version: 6.32.02
Built for linuxx8664gcc on Sep 18 2024, 20:01:03
From heads/master@tags/v6-32-02
Hello ROOT experts. I have a bunch of TH2 histograms representing signal mass distributions binned in the space of two invariant masses (let’s say m_X and m_Y). For a given m_Y, I would like to generate a new signal shape at X mass m_x by interpolating between the shapes of two other signals at X masses m_{x1} and m_{x2}, for m_{x1} < m_x < m_{x2}. I found several similar threads on the ROOT forum which make use of moment morphing:
- Using RooMomentMorphFuncND
- 2D interpolation using moment morphing - #3 by diboye
- [RF] Problem writing RooMomentMorphND to workspace in RooFit · Issue #11876 · root-project/root · GitHub
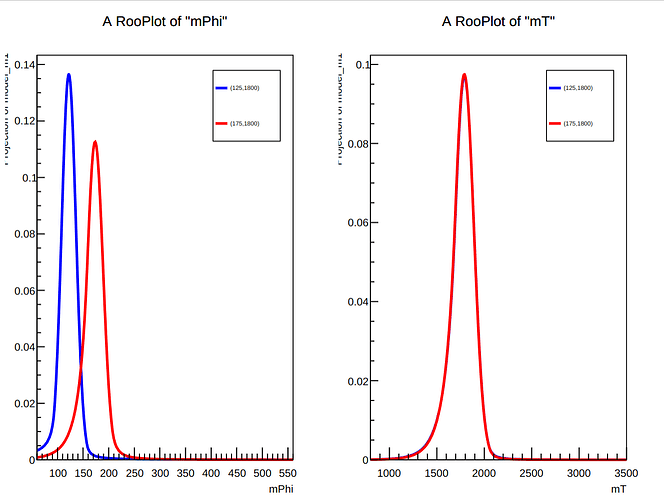
In particular, I verified that I could reproduce the results in the example code from the first link above.
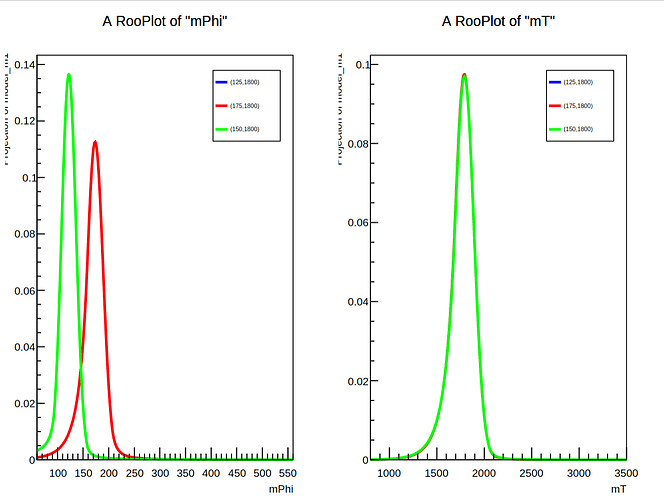
I’ve written a similar function based on the linked code and attached it in the file Interpolator.py. This file opens two (attached) ROOT files and gets the TH2 histograms for signals of m_Y=1800 and m_{x1} = 125 and m_{x2} = 175. The code then creates a RooDataHist for each distribution which is used to make a RooHistPdf. I then create a RooMomentMorphFuncND.Grid2 object to which I add the two PDFs and then attempt to perform linear interpolation between them at the given mass point (150, 1800) using RooMomentMorphFuncND.Linear. However, the code fails with the error:
# DEBUG----------------------------------------------------------------------------------
RooRealVar::mPhi = 310 L(60 - 560)
<class cppyy.gbl.RooRealVar at 0x5a1c3ee41dd0>
TH1: 68.97920879694776
RDH: 68.97920879694763
RDH (true): 68979.20879694764
RHP: 0.0
TH1: 64.96953476283132
RDH: 64.96953476283137
RDH (true): 64969.534762831376
RHP: 5.955384569195107e-06
# DEBUG----------------------------------------------------------------------------------
[#0] ERROR:InputArguments -- Attempting to integrate the RooRealVar "mPhi", but integrating a RooAbsRealLValue is not allowed!
Traceback (most recent call last):
File "/home/amitav/work/CMS/Tprime/Interpolation/Interpolator.py", line 81, in <module>
Interpolate()
File "/home/amitav/work/CMS/Tprime/Interpolation/Interpolator.py", line 69, in Interpolate
pdf.plotOn(framex, RooFit.LineColor(ROOT.kBlue))
File "/home/amitav/miniconda3/envs/ROOT/lib/python3.12/site-packages/ROOT/_pythonization/_roofit/_rooabspdf.py", line 79, in plotOn
return self._plotOn(*args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
cppyy.gbl.std.runtime_error: RooPlot* RooAbsPdf::plotOn(RooPlot* frame, const RooCmdArg& arg1 = {}, const RooCmdArg& arg2 = {}, const RooCmdArg& arg3 = {}, const RooCmdArg& arg4 = {}, const RooCmdArg& arg5 = {}, const RooCmdArg& arg6 = {}, const RooCmdArg& arg7 = {}, const RooCmdArg& arg8 = {}, const RooCmdArg& arg9 = {}, const RooCmdArg& arg10 = {}) =>
runtime_error: Attempting to integrate the RooRealVar "mPhi", but integrating a RooAbsRealLValue is not allowed!
I understand what the error message is saying, but it’s not clear to me why I see this failure mode when the same RooRealVar objects worked in the example code. Am I making a mistake in my code somewhere, perhaps where I convert the RooDataHist to a RooHistPdf? The RooHistPdf.analyticalIntegral(0) call returning 0 doesn’t seem correct to me.
Any help would be much appreciated! I’ve attached a minimal reproducible example to this post as well. Thanks in advance ![]()
Interpolator.py (2.5 KB)
THselection_TprimeB-1800-125_18.root (630.4 KB)
THselection_TprimeB-1800-175_18.root (746.7 KB)
The code is attached in the file Interpolator.py, but included here as well:
import ROOT
from ROOT import TFile, RooRealVar, RooArgList, RooDataHist, RooHistPdf, RooFit, RooArgSet, RooMomentMorphFuncND, RooBinning, RooWrapperPdf
# X = mPhi
xMin = 60
xMax = 560
nX = 50
# Y = mT
yMin = 800
yMax = 3500
nY = 27
def create_RDH_PDF(name, h):
xvar = RooRealVar('mPhi','mPhi',xMin,xMax)
yvar = RooRealVar('mT','mT',yMin,yMax)
RAL_myvars = RooArgList(xvar,yvar)
RAS_myvars = RooArgSet(RAL_myvars)
RDH = RooDataHist('RDH','RDH',RAL_myvars,h)
RHP = RooHistPdf('RHP','RHP',RAS_myvars,RDH)
# DEBUG
print('TH1: ' + str(h.Integral()))
print('RDH: ' + str(RDH.sum(False)))
print('RDH (true): ' + str(RDH.sum(True)))
print('RHP: ' + str(RHP.analyticalIntegral(0)))
return RDH, RHP
def getBin(binning, val):
if (val > binning.highBound()):
return binning.numBins()
else:
return binning.binNumber(val)
def Interpolate():
# Observables
x = RooRealVar('mPhi','mPhi',xMin,xMax)
y = RooRealVar('mT','mT',yMin,yMax)
print(x, type(x))
# Interpolated observables
ix = RooRealVar('i_mPhi','i_mPhi',xMin,xMax)
iy = RooRealVar('i_mT','i_mT',yMin,yMax)
# Interpolated binning
RBx = RooBinning(nX,xMin,xMax)
RBy = RooBinning(nY,yMin,yMax)
grid = RooMomentMorphFuncND.Grid2(RBx,RBy)
# Get the PDFs for the two masses we want to interpolate b/w
f1 = TFile.Open('THselection_TprimeB-1800-125_18.root')
f2 = TFile.Open('THselection_TprimeB-1800-175_18.root')
h1 = f1.Get('MHvsMTH_SR_pass__nominal')
h2 = f2.Get('MHvsMTH_SR_pass__nominal')
RDH1, RHP1 = create_RDH_PDF('1',h1)
RDH2, RHP2 = create_RDH_PDF('2',h2)
# Add the PDFs to the grid at their X,Y location
grid.addPdf(RHP1, getBin(RBx, 1800), getBin(RBy, 125))
grid.addPdf(RHP2, getBin(RBx, 1800), getBin(RBy, 175))
# Morph
morph = RooMomentMorphFuncND('morph', 'morph', RooArgList(ix,iy),RooArgList(x,y), grid, RooMomentMorphFuncND.Linear)
morph.setPdfMode()
pdf = RooWrapperPdf('morph_pdf', 'morph_pdf', morph, True)
# Get the frames for plotting
framex = x.frame()
framey = y.frame()
# Generate at a given point
ix.setVal(1800)
ix.setVal(150)
# Plot
pdf.plotOn(framex, RooFit.LineColor(ROOT.kBlue))
pdf.plotOn(framey, RooFit.LineColor(ROOT.kBlue))
c = ROOT.TCanvas('c','c')
c.Divide(2,1)
c.cd(1)
framex.Draw()
c.cd(2)
framey.Draw()
framey.Draw('same')
c.Print('out.pdf')
Interpolate()