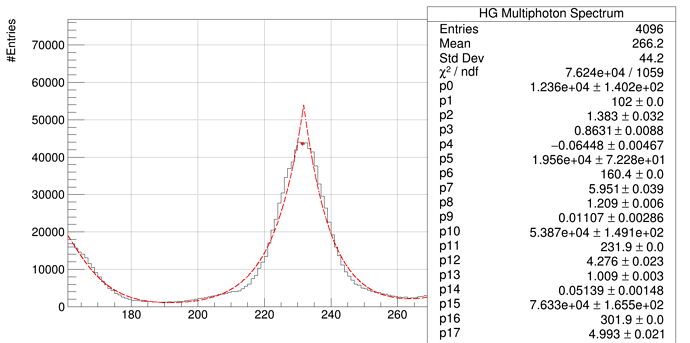
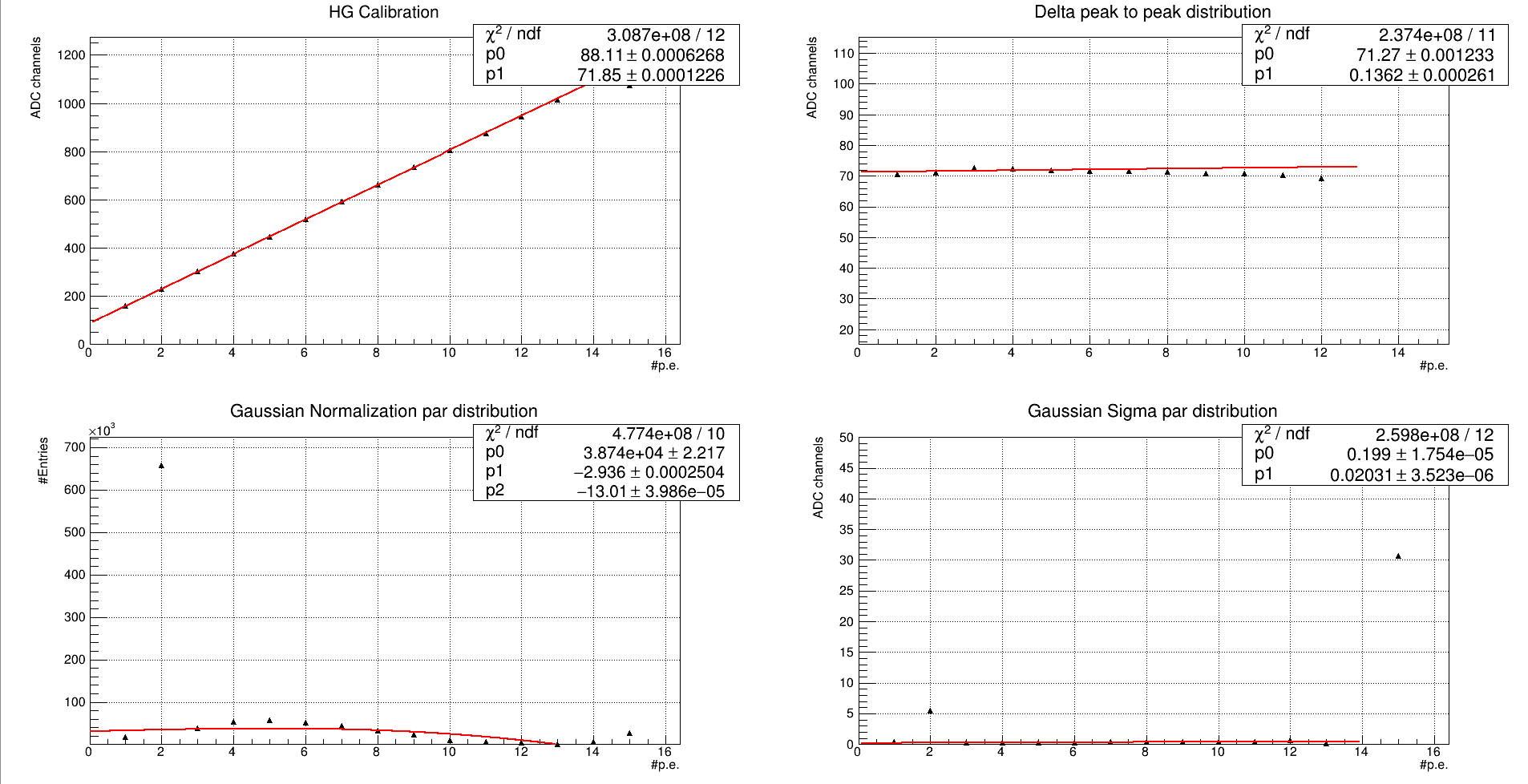
Hi ROOTers,
We wrote this code in order to analize multiphoton spectrum:
FILE* file;
TH1F* hsipm;
float x;
int bins = 0;
TF1* full;
int sigma = 5;
const Int_t npeaks = 40;
Double_t par[3*npeaks];
Double_t fpeaks(Double_t *x, Double_t *par) {
Double_t result = 0.;
for (Int_t p=0;p<npeaks;p++) {
Double_t norm = par[3*p+0];
Double_t mean = par[3*p+1];
Double_t sigma = par[3*p+2];
result += norm*TMath::Gaus(x[0],mean,sigma);
}
return result;
}
Double_t gain = 0.;
Double_t pedestal = 0.;
/*
//par[0]=mu - par[1]=p - par[2]=N (articolo vinogradov sipm1 pag. 6)
Double_t Vino(Double_t* x, Double_t* par){
Double_t mu = par[0];
Double_t p = par[1];
//Double_t N = par[2];
Int_t N = int((x[0]-pedestal)/gain);
Double_t B;
Double_t f0 = exp(-mu);
Double_t term = mu*(1-p);
Double_t sum = 0.;
if(N==0)
return f0;
for(int i=1; i<N; ++i){
B = TMath::Gamma(N)/(TMath::Gamma(i+1)*TMath::Gamma(i)*TMath::Gamma(N-i+1));
sum += B*pow(term,i)*pow(p,N-i);
}
return sum*f0;
}
*/
int Multiphoton_spectrum_analysis() {
file = fopen("Run0_PHA_HG_0_10.txt", "r");
TCanvas* c1 = new TCanvas("c1","HG Multiphoton Spectrum",10,10,1920,1980);
gStyle->SetOptFit(000);
c1->SetGridx();
c1->SetGridy();
TH1F* htmp = new TH1F("htemp","htemp",4096,0,4096);
while(!feof(file)){
fscanf(file,"%g\n",&x);
htmp->SetBinContent(bins,x);
++bins;
}
hsipm = new TH1F("HG Multiphoton Spectrum","",bins,0,bins);
for(int a=0; a<bins; ++a)
hsipm->SetBinContent(a,htmp->GetBinContent(a));
sigma = bins/1024+1;
hsipm->GetYaxis()->SetTitle("#Entries");
hsipm->GetXaxis()->SetTitle("ADC channels");;
hsipm->GetXaxis()->SetRange(0,1300);
TF1* f = new TF1("f",fpeaks,0,4096,3*npeaks);
f->SetParameters(par);
TSpectrum *s = new TSpectrum(npeaks);
Int_t nfound;
TString peak_name;
TString full_func = "";
TF1* g[npeaks];
nfound = s->Search(hsipm,sigma,"",0.0007);
printf("Found %d candidate peaks to fit\n", nfound);
int peak_x_tmp[npeaks];
double peak_x[npeaks];
double peak_y[npeaks];
TMath::Sort(nfound,s->GetPositionX(),peak_x_tmp,kFALSE);
for(int a=0; a<nfound; ++a){
peak_x[a] = s->GetPositionX()[peak_x_tmp[a]];
peak_y[a] = s->GetPositionY()[peak_x_tmp[a]];
}
for(int a=0; a<nfound; ++a){
peak_name.Form("g%d",a);
g[a] = new TF1(peak_name,"gaus");
TString func_tmp;
func_tmp.Form("gaus(%d)+",3*a);
full_func += func_tmp;
}
full_func.Remove(full_func.Length()-1);
full = new TF1("full",full_func);
for(int a=2; a<nfound; ++a){
full->SetParLimits(3*a+0,peak_y[a]*0.5,peak_y[a]*1.1);
full->SetParLimits(3*a+1,peak_x[a]-3*sigma, peak_x[a]+3*sigma);
full->SetParLimits(3*a+2,0.5*sigma,6*sigma);
}
// pedestal
full->SetParLimits(0,peak_y[0]*0.6,peak_y[0]*1.1);
full->SetParLimits(1,peak_x[0]-1.2*sigma, peak_x[0]+1.2*sigma);
full->SetParLimits(2,0.5*sigma,1*sigma);
// 1 p.e.
full->SetParLimits(3,peak_y[1]*0.5,peak_y[1]*1.1);
full->SetParLimits(4,peak_x[1]-1.5*sigma, peak_x[1]+1.5*sigma);
full->SetParLimits(5,0.5*sigma,4*sigma);
full->SetNpx(4096*2);
full->SetLineStyle(9);
full->SetLineColor(2); //colore fit generale
hsipm->Fit("full","","same",35.,peak_x[nfound-1]+4*sigma);
hsipm->SetLineColor(1); //colore dei dati
double fPar[npeaks] = {0.};
double fErr[npeaks*3] = {0.};
full->GetParameters(&fPar[0]);
for(int a=0; a<nfound*3; ++a)
fErr[a] = full->GetParErrors()[a];
TF1* fg[npeaks];
TString gname;
double m_tmp = fPar[1];
double s_tmp = fPar[2];
fg[0] = new TF1("ped","gaus",m_tmp-3*s_tmp,m_tmp+3*s_tmp);
fg[0]->SetParameters(&fPar[0]);
fg[0]->SetLineColor(9); //colore piedistallo
//fg[0]->Draw("same");
for(int a=1; a<nfound; ++a){
m_tmp = fPar[3*a+1];
s_tmp = fPar[3*a+2];
gname.Form("%d p.e.",a);
fg[a] = new TF1(gname,"gaus",m_tmp-3*s_tmp,m_tmp+3*s_tmp);
fg[a]->SetParameters(&fPar[3*a]);
fg[a]->SetLineColor(9); //colore gaussiane
//fg[a]->Draw("same");
}
double aNpeak[npeaks];
double aConst[npeaks];
double aMean[npeaks];
double aSig[npeaks];
double aConstErr[npeaks];
double aMeanErr[npeaks];
double aSigErr[npeaks];
const double sqrt2pi = sqrt(2 * acos(-1.));
for (int a = 1; a < nfound; ++a)
{
aNpeak[a - 1] = a;
aMean[a - 1] = fPar[3 * a + 1];
aMeanErr[a - 1] = fErr[3 * a + 1];
aSig[a - 1] = fPar[3 * a + 2];
aSigErr[a - 1] = fErr[3 * a + 2];
aConst[a - 1] = fPar[3 * a] * sqrt2pi * aSig[a - 1];
aConstErr[a - 1] = sqrt2pi * (fErr[3 * a] * aSig[a - 1] + aSigErr[a - 1] * fPar[3 * a]);
}
FILE* outfile;
outfile = fopen("Gaussian_Parameters.txt", "w");
for(int a=0; a<nfound; ++a)
fprintf(outfile, "%d \t %f \t %f \t %f \t %f \t %f \t %f \n", a, fPar[3 * a], fErr[3*a], fPar[3 * a + 1], fErr[3*a+1], fPar[3 * a + 2], fErr[3*a+2]);
fclose(outfile);
TCanvas* c2 = new TCanvas("c2","HG Multiphoton Spectrum Analysis",10,10,1920,1980);
c2->UseCurrentStyle();
gStyle->SetOptFit(111);
c2->Divide(2,2);
c2->cd(1);
c2->GetPad(1)->SetGridx();
c2->GetPad(1)->SetGridy();
c2->Update();
Double_t xErr[npeaks] = {0.};
TGraphErrors* mean = new TGraphErrors(nfound-1,aNpeak,aMean,xErr,aMeanErr);
mean->SetMarkerStyle(22);
mean->SetTitle("HG Calibration");
mean->GetXaxis()->SetTitle("#p.e.");
mean->GetYaxis()->SetTitle("ADC channels");
mean->Draw("ap");
TF1* fgain = new TF1("fgain","pol1");
mean->Fit("fgain","","same",0.,nfound-2);
mean->GetYaxis()->SetRangeUser(0.,aMean[nfound-2]+200.);
c2->cd(1)->Update();
pedestal = fgain->GetParameter(0);
gain = fgain->GetParameter(1);
c2->cd(2);
c2->GetPad(2)->SetGridx();
c2->GetPad(2)->SetGridy();
Double_t aDpp[npeaks] = {0.};
Double_t aDppErr[npeaks] = {0.};
Double_t mean_dpp = 0.;
for(int a=0; a<nfound-2; ++a){
aDpp[a] = aMean[a+1]-aMean[a];
aDppErr[a] = aMeanErr[a+1]+aMeanErr[a];
mean_dpp += aDpp[a];
}
mean_dpp /= (nfound-2);
TGraphErrors* gDpp = new TGraphErrors(nfound-2,aNpeak,aDpp,xErr,aDppErr);
gDpp->SetMarkerStyle(22);
gDpp->SetTitle("Delta peak to peak distribution");
gDpp->GetXaxis()->SetTitle("#p.e.");
gDpp->GetYaxis()->SetTitle("ADC channels");
gDpp->Draw("ap");
gDpp->Fit("pol1","","same",0.,nfound-3);
gDpp->GetYaxis()->SetRangeUser(mean_dpp-50,mean_dpp+50.);
c2->cd(3);
c2->GetPad(3)->SetGridx();
c2->GetPad(3)->SetGridy();
TGraphErrors* gConst = new TGraphErrors(nfound-1,aNpeak,aConst,xErr,aConstErr);
gConst->SetMarkerStyle(22);
gConst->SetTitle("Gaussian Normalization par distribution");
gConst->GetXaxis()->SetTitle("#p.e.");
gConst->GetYaxis()->SetTitle("#Entries");
gConst->Draw("ap");
TF1 *poisson = new TF1("poisson","[0]*TMath::Power(([1]/[2]),(x/[2]))*(TMath::Exp(-([1]/[2])))/TMath::Gamma((x/[2])+1.)", 0, nfound);
poisson->SetParameters(1.e6, 9., 2.);
poisson->SetNpx(4096*2);
gConst->Fit(poisson,"R","same",0.,nfound-2);
c2->cd(4);
c2->GetPad(4)->SetGridx();
c2->GetPad(4)->SetGridy();
TGraphErrors* gSig = new TGraphErrors(nfound-1,aNpeak,aSig,xErr,aSigErr);
gSig->SetMarkerStyle(22);
gSig->SetTitle("Gaussian Sigma par distribution");
gSig->GetXaxis()->SetTitle("#p.e.");
gSig->GetYaxis()->SetTitle("ADC channels");
gSig->Draw("ap");
gSig->GetYaxis()->SetRangeUser(0.,50.);
TF1* pol1 = new TF1("pol1","pol1");
pol1->SetParameters(20.,0.5);
gSig->Fit("pol1","","same",0.,nfound-2);
return 0;
}
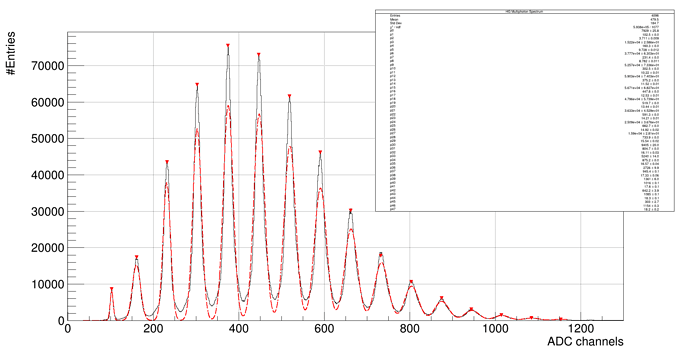
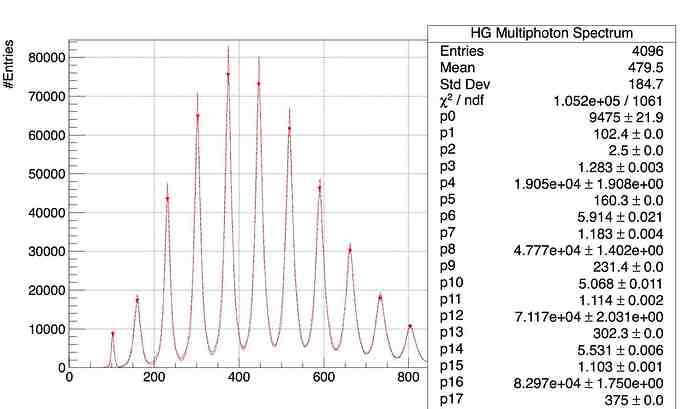
And we get this results:
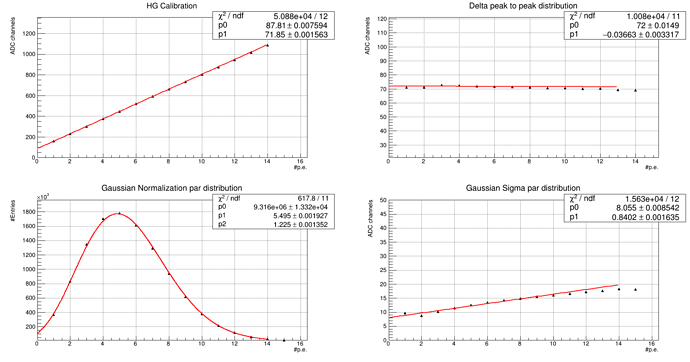
and
meanwhile in the terminal i get this warning:
Info in <ROOT::Math::ParameterSettings>: lower/upper bounds outside current parameter value. The value will be set to (low+up)/2
-
How can I delete this warning? I suppose that i need to set parameterlimits but i did it.
-
Why the chi squared are so big? The points lie on the lines…
-
I saw that TSpectrum is deprecated, is there an alternative?
-
There is a way to set two different style about statistic table? I used
gStyle->SetOptFit(000)andgStyle->SetOptFit(111)in the two canvas but doesnt work. -
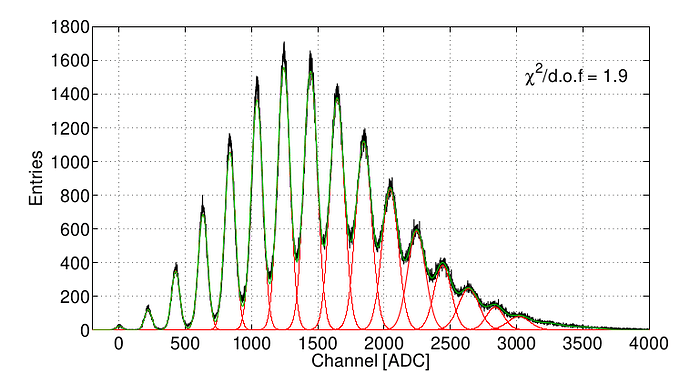
There is a way to do a similar fit (green) in ROOT? How?
I mean, do a multi gaussian fit like green line.