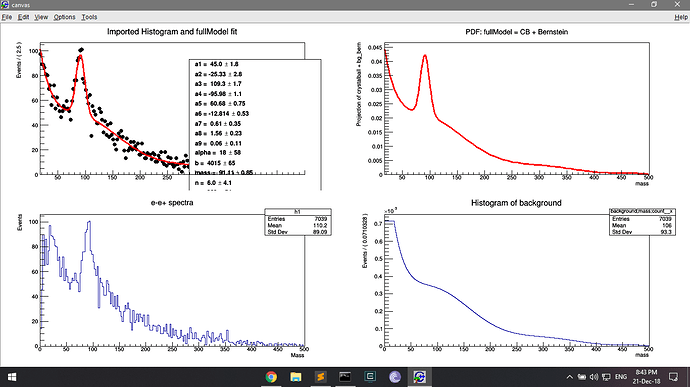
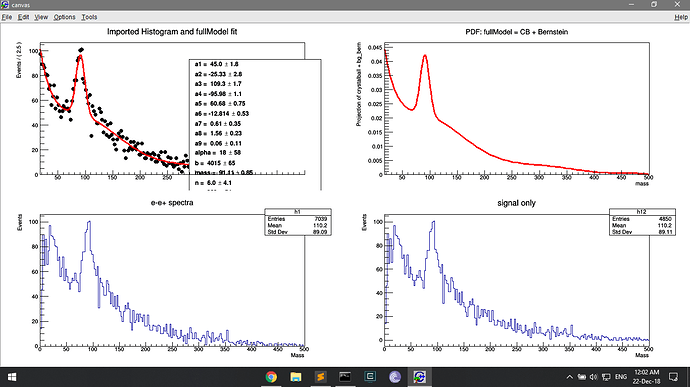
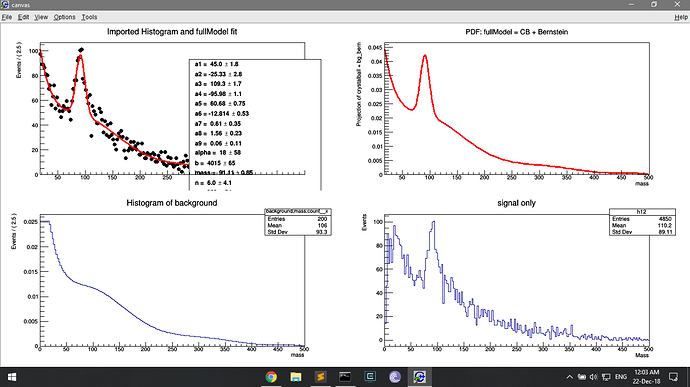
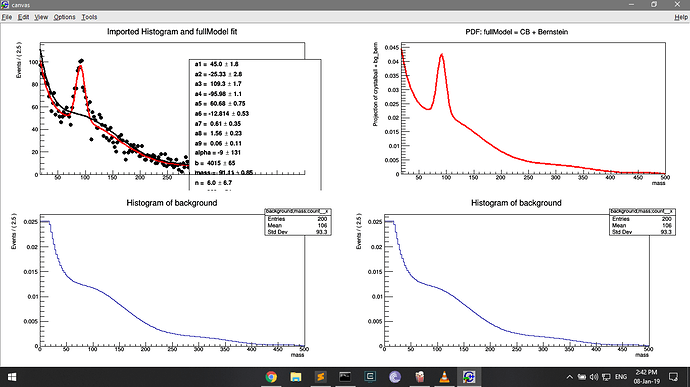
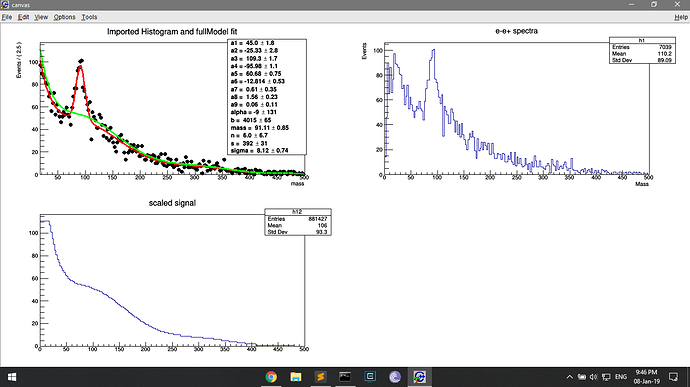
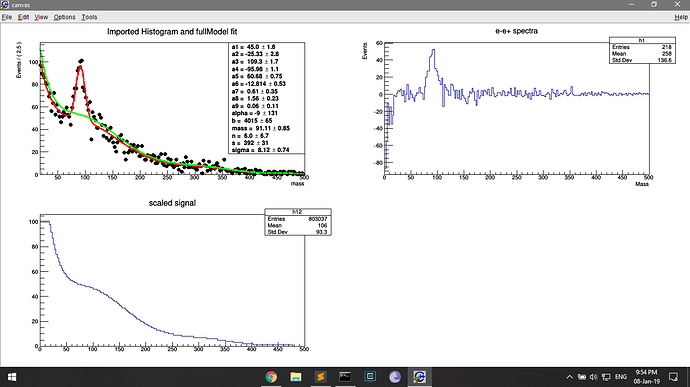
I think the problem starts with my roopdf. I am trying to fit the h1 histo.
TH1F *h1 = new TH1F("h1","e-e+ spectra;Mass;Events",200,0.,500.);
RooDataHist dh("dh","e-e+",x,Import(*h1)) ;
the dh histo has been drawn on the first panel and it seems to retain the count of the original histo (h1).
I am then fitting it with a pdf fullmodel
RooRealVar x("x","mass",18.,500.);
x.setRange("Range1",18.,75.) ;
x.setRange("Range2",107.,495.) ;
RooRealVar a1("a1","a1",-50,1000);
RooRealVar a2("a2","a2",-50,400);
RooRealVar a3("a3","a3",-50,400);
RooRealVar a4("a4","a4",-500,1000);
RooRealVar a5("a5","a5",-50,1000);
RooRealVar a6("a6","a6",-50,1000);
RooRealVar a7("a7","a7",-50,1000);
RooRealVar a8("a8","a8",-50,100);
RooRealVar a9("a9","a9",-50,100);
RooRealVar aa("aa","aa",-50,100);
RooRealVar mass("mass","Central value of Gaussian",90.,80.,100.);
RooRealVar sigma("sigma","Width of Gaussian",20,0,100);
RooRealVar alpha("alpha","Alpha",40.,0.,100.);
RooRealVar n("n","Order",6,0.,10.);
RooCBShape crystalball("crystalball", "Signal Region", x,mass,sigma,alpha,n);
RooRealVar b("b", "Number of background events", 0, 5500);
RooRealVar s("s", "Number of signal events", 0, 500);
RooBernstein bg_bern("bg_bern","background",x, RooArgList(a1,a2,a3,a4,a5,a6,a7,a8,a9));
RooAddPdf fullModel("fullModel", "crystalball + bg_bern", RooArgList(crystalball, bg_bern),RooArgList(s, b));
RooFitResult* r = fullModel.fitTo(dh,Save()) ;
But drawing this pdf on the second panel shows the low counts (because normalised?). This is where I am deriving the bg_bern pdf, and this must be where the low count issue arises.
TH1 *h2 = bg_bern.createHistogram("background;mass;count", x, Binning(200,0.,500.));