Dear experts,
I am trying to perform a fit to a sWeighted data, then do second sWeight calculation based on my new fit result.
The following is my workflow.
First, I use a binned fit to extract the distribution of signal and background. The binned fit is employed because of the large amount of data ( ~10^6 events originally).
Blockquote
//input original root file
TFile *input_root_1 = new TFile(filename);
TTree tree1 = (TTree)input_root_1->Get(“DecayTree”);
// input root file which stores the sWeighted factor
TFile *input_root_2 = new TFile(sweight_file);
TTree tree2 = (TTree)input_root_2->Get(“DecayTree”);
//add tree friend
tree1->AddFriend(tree2);
//fit variable
RooRealVar ipchi(“log_D_IPCHI2_OWNPV”, “ipchi”, xmin, xmax);
//sweight factor
TString weight(“sig_sw”);
//fit range
Double_t xmin=-6.5, xmax=8.5; Double_t xbins=75;
// create RooDataHist
TH1F *th1f_type1=new TH1F(“th1f_type1”,“th1f_type1”, xbins, xmin, xmax);
tree1->Project(“th1f_type1”,draw_var, weight);
RooDataHist data(“data”,“ipchi”,ipchi, th1f_type1);
//event number of data
double N = data.sumEntries();
…
//signal distribution
RooRealVar* ap_p;
RooRealVar* sigp_p;
RooRealVar* xi_p;
RooRealVar* rho1_p;
RooRealVar* rho2_p;
TFile* input_ws_file = new TFile(“ws.roott”);
RooWorkspace* w = (RooWorkspace*) input_ws_file->Get(“w”) ;
ap_p = w->var(“ap_p”);
sigp_p = w->var(“sigp_p”);
xi_p = w->var(“xi_p”);
rho1_p = w->var(“rho1_p”);
rho2_p = w->var(“rho2_p”);
ap_p->setConstant();
rho2_p->setConstant();
//sig pdf
RooBukinPdf sgn_p(“sgn_p”,“sgn_p”,ipchi,*ap_p,*sigp_p,*xi_p,rho1_p,rho2_p);
//sig number
RooRealVar nsig_prompt(“nsig_prompt”, “nsig_prompt”, 0.8N, 0.6N, N);
//background
RooRealVar bdecay_mean(“bdecay_mean”,“bdecay_mean”, 4.5, 4.0, 5.0);
RooRealVar bdecay_sigma(“bdecay_sigma”, “bdecay_sigma”, 1.2, 0.8, 2.0);
//bkg pdf
RooGaussian sgn_b(“sgn_b”, “sgn_b”, ipchi, bdecay_mean, bdecay_sigma);
//bkg number
RooRealVar nsig_bdecay(“nsig_bdecay”, “nsig_bdecay”, 0.2N, 0, 0.3N);
// total pdf
RooArgList shapes;
RooArgList yields;
shapes.add(sgn_p); yields.add(nsig_prompt);
shapes.add(sgn_b); yields.add(nsig_bdecay);
RooAddPdf *model = new RooAddPdf(“model”, “sum of all PDFs”, shapes, yields);
// binned fit
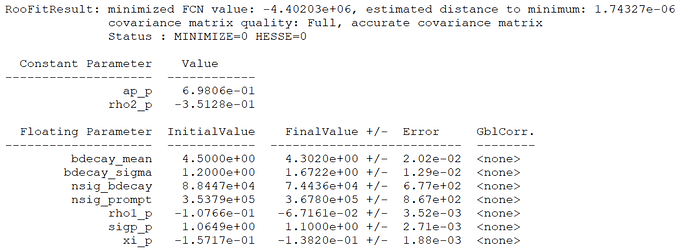
m_fitres = model->fitTo(data, NumCPU(40), Save(kTRUE), Extended(kTRUE));
m_fitres->Print(“v”);
Blockquote
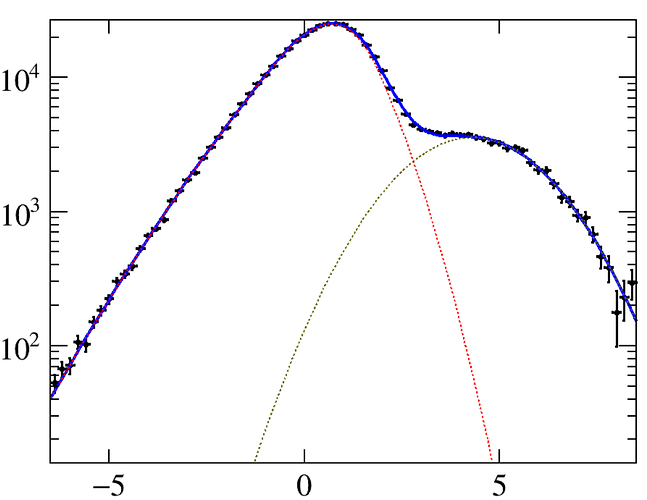
The fit quality is good, as can be seen here.
Next, the RooDataSet is used in order to calculate sWeight factor.
Blockquote
// create sWeighted RooDataSet
RooRealVar* mass_weight = new RooRealVar(“sig_sw”, “sig_sw”, -5, 5);
RooDataSet data_sw(“data_sw”, “ipchi”, tree1, RooArgSet(ipchi,*mass_weight),0, mass_weight->GetName());
//SPlot
RooStats::SPlot* sData = new RooStats::SPlot(“sData”,“An SPlot”, data_sw, model, RooArgList(nsig_prompt, nsig_bdecay));
Blockquote
This plot displays the input RooDataSet and model. It doesn’t look like there’s anything wrong with it.
But I have encounter a problem when caluculating sWeight factor. An Error reminder shows
and it results in fail sWeight calculation.
I know this problem I’m having has been asked before, and in its answer, simply changing the fit parameter floating range can solve it. But I have tried many times and it does not solve the problem.
How can I fix it?
Boan
(Previous discussions)
https://root-forum.cern.ch/t/error-eval-rooabsreal-logevalerror-xxx-evaluation-error/16228