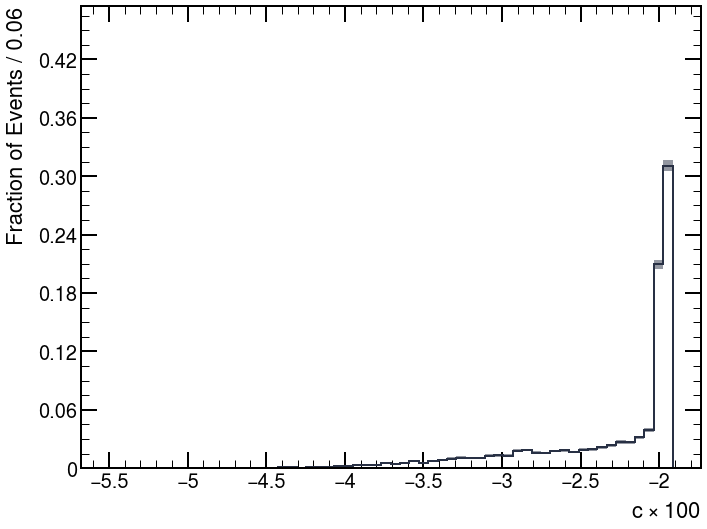
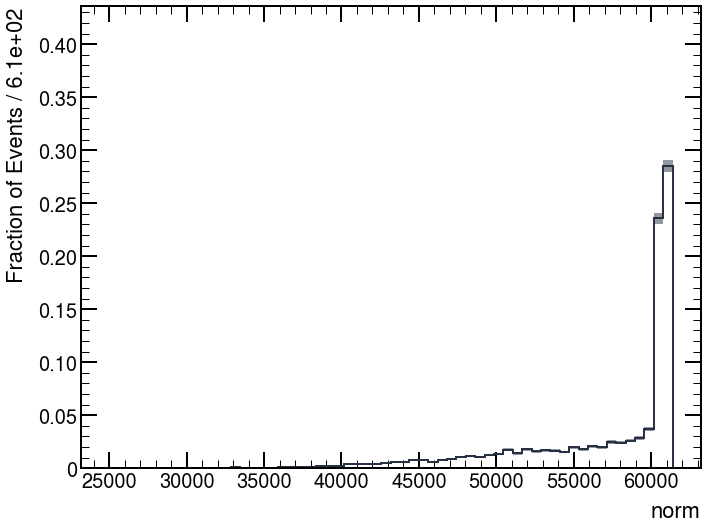
Hi @jonas ,
Concerning 1: I did use the cholesky matrix in my implementation
Concerning 2: I still think the boundary should be adjusted. Please see my reproducer below. For the x -> t correction, after more careful inspection, I think the correction should instead be (high - nominal) -> (high - nominal) / boundary and (nominal - low) -> (nominal - low) / boundary
Please see below my reproducer
from typing import Optional, Union
import numpy as np
import ROOT
observable = ROOT.RooRealVar('myy', 'myy', 125, 105, 160)
observable.setBins(55)
c = ROOT.RooRealVar('c', 'c', 1, -10, 10)
base_pdf = ROOT.RooExponential('exp', 'exp', observable, c)
norm = ROOT.RooRealVar('N', 'N', 1, -float('inf'), float('inf'))
pdf = ROOT.RooAddPdf('exp_extended', '', ROOT.RooArgList(base_pdf), ROOT.RooArgList(norm))
observable.setRange('SB_Lo', 105, 120)
observable.setRange('SB_Hi', 130, 160)
bin_edges = np.linspace(105, 160, 55 + 1)
x = (bin_edges[:-1] + bin_edges[1:]) / 2
y = np.array([1201.58, 1179.01, 1140.39, 1128.87, 1111.56, 1124.37, 1098.3 ,
1052.13, 1015.04, 1006.18, 971.46, 945.95, 931.43, 913.02,
871.57, 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 724.1 , 699.51, 687.49,
667.44, 657.39, 649.17, 634.78, 600.47, 586.51, 580.54,
570.01, 588.22, 555.9 , 540.72, 531.02, 546.06, 528.99,
521.36, 495.24, 483.4 , 448.63, 480.71, 454.2 , 450.19,
436.4 , 413.69, 412.14, 420.22, 417.97, 404.38])
arrays = {
'myy': x,
'weight': y
}
dataset = ROOT.RooDataSet.from_numpy(arrays, ROOT.RooArgSet(observable), weight_name='weight')
fit_args = [ROOT.RooFit.Range('SB_Lo,SB_Hi'), ROOT.RooFit.PrintLevel(-1),
ROOT.RooFit.Hesse(True), ROOT.RooFit.Save(), ROOT.RooFit.Strategy(2),
ROOT.RooFit.AsymptoticError(True)]
fit_result = pdf.fitTo(dataset, *fit_args)
def get_covariance_matrix(fit_result):
data = fit_result.covarianceMatrix()
array = data.GetMatrixArray()
nrows = data.GetNrows()
ncols = data.GetNcols()
cov_matrix = np.frombuffer(array, dtype='float64', count=nrows * ncols).reshape((nrows, ncols))
return cov_matrix
# covariance matrix
C = get_covariance_matrix(fit_result)
# > print(C)
# > [[ 4.11727600e+07 -2.15903749e+01]
# > [-2.15903749e+01 8.21503626e-05]]
# cholesky matrix
# here L is same as _Lt in https://github.com/root-project/root/blob/master/roofit/roofitcore/src/RooFitResult.cxx#L365
L = np.linalg.cholesky(C)
# > print(L)
# > [[ 6.41660034e+03 0.00000000e+00]
# > [-3.36476853e-03 8.41597857e-03]]
# Sample Gaussian vector, shape = (n_sample, n_param)
np.random.seed(1234)
G = np.random.normal(size=(10, 2))
# > print(G)
# > [[ 4.71435164e-01 -1.19097569e+00]
# > [ 1.43270697e+00 -3.12651896e-01]
# > [-7.20588733e-01 8.87162940e-01]
# > [ 8.59588414e-01 -6.36523504e-01]
# > [ 1.56963721e-02 -2.24268495e+00]
# > [ 1.15003572e+00 9.91946022e-01]
# > [ 9.53324128e-01 -2.02125482e+00]
# > [-3.34077366e-01 2.11836468e-03]
# > [ 4.05453412e-01 2.89091941e-01]
# > [ 1.32115819e+00 -1.54690555e+00]]
# according to https://root.cern.ch/doc/master/TVectorT_8cxx_source.html#l00990
# the line g*= (*_Lt) : https://github.com/root-project/root/blob/master/roofit/roofitcore/src/RooFitResult.cxx#L376
# will call cblas_dgemv, which does _Lt * g
# if we want to multiply multiple vectors G, it's equivalent to doing
# G * _Lt.T -> G @ L.T in our notation
R = G @ L.T
# > print(R)
# > [[ 3.02501103e+03 -1.16094961e-02]
# > [ 9.19310803e+03 -7.45199898e-03]
# > [-4.62372992e+03 9.89095859e-03]
# > [ 5.51563531e+03 -8.24928422e-03]
# > [ 1.00717347e+02 -1.89272032e-02]
# > [ 7.37931963e+03 4.47859246e-03]
# > [ 6.11709993e+03 -2.02185523e-02]
# > [-2.14364094e+03 1.14192112e-03]
# > [ 2.60163250e+03 1.06873470e-03]
# > [ 8.47734411e+03 -1.74641155e-02]]
# note the one-sigma range of this R is definitely not (-1, 1)
parameters = [param for param in fit_result.floatParsFinal()]
# nominal value of parameters
V0 = np.array([param.getVal() for param in parameters])
# > print(V0)
# > [ 3.99293343e+04 -2.06860695e-02]
Err = np.array([[param.getErrorLo(), param.getErrorHi()] for param in parameters])
# > print(Err)
# > [[-6.41660034e+03 2.23800563e+02]
# > [-9.06368372e-03 3.27688567e-04]]
# if I don't do any interpolation:
V_no_interp = V0 + R
# > print(V_no_interp)
# > [[ 4.29543454e+04 -3.22955657e-02]
# > [ 4.91224424e+04 -2.81380685e-02]
# > [ 3.53056044e+04 -1.07951109e-02]
# > [ 4.54449697e+04 -2.89353537e-02]
# > [ 4.00300517e+04 -3.96132727e-02]
# > [ 4.73086540e+04 -1.62074771e-02]
# > [ 4.60464343e+04 -4.09046218e-02]
# > [ 3.77856934e+04 -1.95441484e-02]
# > [ 4.25309668e+04 -1.96173348e-02]
# > [ 4.84066784e+04 -3.81501850e-02]]
# will be roughly gaussian with the correct sigma and mean
# now do the interpolation
# implementation according to https://root.cern/doc/master/MathFuncs_8h_source.html#l00213
# with some correction:
# (high - nominal) -> (high - nominal) / boundary
# (nominal - low) -> (nominal - low) / boundary
def additive_polynomial_linear_extrapolation(
x: Union[float, np.ndarray], nominal: float, low: float, high: float, boundary: float,
):
x = np.asarray(x, dtype=np.float64)
mod = np.zeros_like(x)
mask1 = x >= boundary
mask2 = x <= -boundary
mask3 = ~(mask1 | mask2)
mod[mask1] = x[mask1] * (high - nominal) / boundary
mod[mask2] = x[mask2] * (nominal - low) / boundary
if np.any(mask3):
t = x[mask3] / boundary
eps_plus = (high - nominal) / boundary
eps_minus = (nominal - low) / boundary
S = 0.5 * (eps_plus + eps_minus)
A = 0.0625 * (eps_plus - eps_minus)
mod[mask3] = x[mask3] * (S + t * A * (15 + t**2 * (-10 + t**2 * 3)))
return mod
# case 1: set boundary to the default value of 1
R_trans_boudnary_1 = np.zeros_like(R)
for i, param in enumerate(parameters):
nominal = param.getVal()
low = nominal + param.getErrorLo()
high = nominal + param.getErrorHi()
boundary = 1.0
R_trans_boudnary_1[:, i] = additive_polynomial_linear_extrapolation(R[:, i], nominal=nominal, low=low, high=high, boundary=boundary)
# > print(R_trans_boudnary_1)
# > [[ 6.76999171e+05 -5.56183018e-05]
# > [ 2.05742275e+06 -3.54470407e-05]
# > [-2.96686270e+07 4.56436535e-05]
# > [ 1.23440229e+06 -3.92933592e-05]
# > [ 2.25405989e+04 -9.18094810e-05]
# > [ 1.65149588e+06 2.08657935e-05]
# > [ 1.36901041e+06 -9.82870506e-05]
# > [-1.37548872e+07 5.35142356e-06]
# > [ 5.82246817e+05 5.00908818e-06]
# > [ 1.89723438e+06 -8.45034077e-05]]
# the scaling of R_trans_boudnary_1 is obviously not correct
# case 2: set boundary to sqrt(C_ii)
R_trans_boudnary_sqrtCii = np.zeros_like(R)
for i, param in enumerate(parameters):
nominal = param.getVal()
low = nominal + param.getErrorLo()
high = nominal + param.getErrorHi()
boundary = np.sqrt(C[i, i])
R_trans_boudnary_sqrtCii[:, i] = additive_polynomial_linear_extrapolation(R[:, i], nominal=nominal, low=low, high=high, boundary=boundary)
# > print(R_trans_boudnary_sqrtCii)
# > [[ 4.53363029e+02 -1.16094961e-02]
# > [ 3.20640626e+02 -7.40801203e-03]
# > [-4.52612470e+03 3.57597876e-04]
# > [ 2.08911318e+02 -8.24255141e-03]
# > [ 5.06849388e+01 -1.89272032e-02]
# > [ 2.57378642e+02 6.22240610e-04]
# > [ 2.14078889e+02 -2.02185523e-02]
# > [-1.71057236e+03 4.62971299e-04]
# > [ 4.91206241e+02 4.40866735e-04]
# > [ 2.95675947e+02 -1.74641155e-02]]
V_interap_boundary_1 = V0 + R_trans_boudnary_1
# > print(V_interap_boundary_1)
# > [[ 7.16928506e+05 -2.07416878e-02]
# > [ 2.09735208e+06 -2.07215166e-02]
# > [-2.96286976e+07 -2.06404259e-02]
# > [ 1.27433162e+06 -2.07253629e-02]
# > [ 6.24699332e+04 -2.07778790e-02]
# > [ 1.69142522e+06 -2.06652037e-02]
# > [ 1.40893974e+06 -2.07843566e-02]
# > [-1.37149579e+07 -2.06807181e-02]
# > [ 6.22176152e+05 -2.06810604e-02]
# > [ 1.93716372e+06 -2.07705729e-02]]
V_interap_boundary_sqrtCii = V0 + R_trans_boudnary_sqrtCii
# > print(V_interap_boundary_sqrtCii)
# > [[ 4.03826974e+04 -3.22955657e-02]
# > [ 4.02499750e+04 -2.80940816e-02]
# > [ 3.54032096e+04 -2.03284717e-02]
# > [ 4.01382457e+04 -2.89286209e-02]
# > [ 3.99800193e+04 -3.96132727e-02]
# > [ 4.01867130e+04 -2.00638289e-02]
# > [ 4.01434132e+04 -4.09046218e-02]
# > [ 3.82187620e+04 -2.02230982e-02]
# > [ 4.04205406e+04 -2.02452028e-02]
# > [ 4.02250103e+04 -3.81501850e-02]]
Now, the corrected distributions become:
Do you think the above implementation is correct?
Thanks,
Alkaid