Dear All,
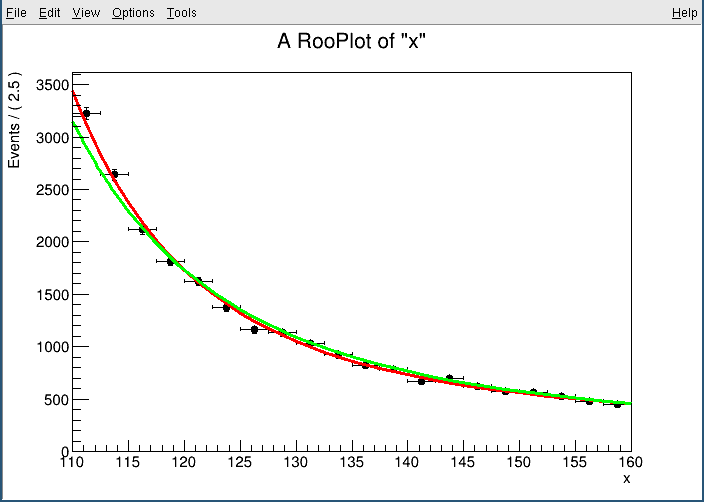
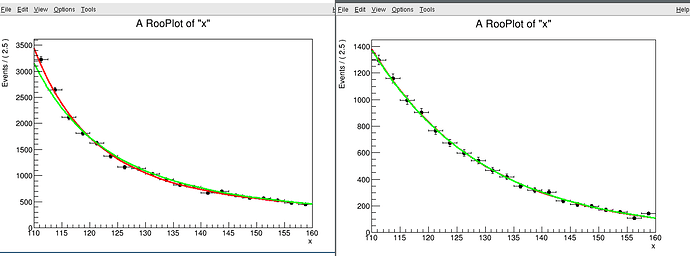
I want to perform a fit in two side-band regions, for example (110,120) and (130,160). As a check, I performed the fit in the side-bands that I expect to be equivalent to the full fit, (110,115)+(115,160) = (110,160):
x.setRange('low',110,115) # low side-band range
x.setRange('high',115,160) # high side-band range
x.setRange('full',110,160) # full range
fitResultFull = pdf.fitTo(data,Range("full"))
fitResultSideBand = pdf.fitTo(data,Range("low,high"))
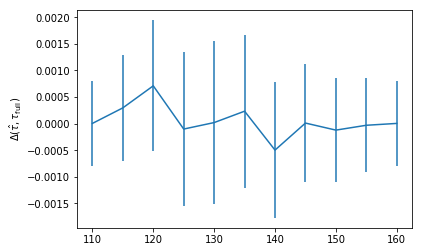
However, for these two fits I get slightly different results. Is there any reason to expect this?
=== Minimal example ===
This is a minimal working example as an illustration using pyroot, python 2.7, ROOT 6.12. The input workspace is attached and saved in save.root
import sys
# please change to point to a python2, root 6.12 pyroot installation
# sys.path = ["path"] + sys.path
import ROOT # using ROOT 6.12
# load from saved file
f=ROOT.TFile("save.root","update")
w=f.Get("w")
# these objects should be in the workspace
bkgName="bkgDijetGgh1"
bkgPdfName="bkgPdfDijetGgh1"
# set up workspace
cut=115
w.var('x').setRange('low',110,cut)
w.var('x').setRange('high',cut,160)
w.var('x').setRange('full',110,160)
x=w.var('x')
# make plot
frame = x.frame()
data=w.obj(bkgName)
data.plotOn(frame)
# full
pdf = w.pdf(bkgPdfName).Clone()
fitResult = pdf.fitTo(data,ROOT.RooFit.Range("full"))
pdf.plotOn(frame,ROOT.RooFit.Normalization(1,ROOT.RooAbsReal.Relative),ROOT.RooFit.LineColor(2))
# part (side band fit)
pdf = w.pdf(bkgPdfName).Clone()
fitResult = pdf.fitTo(data,ROOT.RooFit.Range("low,high"))
pdf.plotOn(frame,ROOT.RooFit.LineColor(3))
# draw plot and keep open
frame.Draw()
raw_input()
save.root (13.7 KB)