Hi,
I have a problem with the integration of my custom PDF classes.
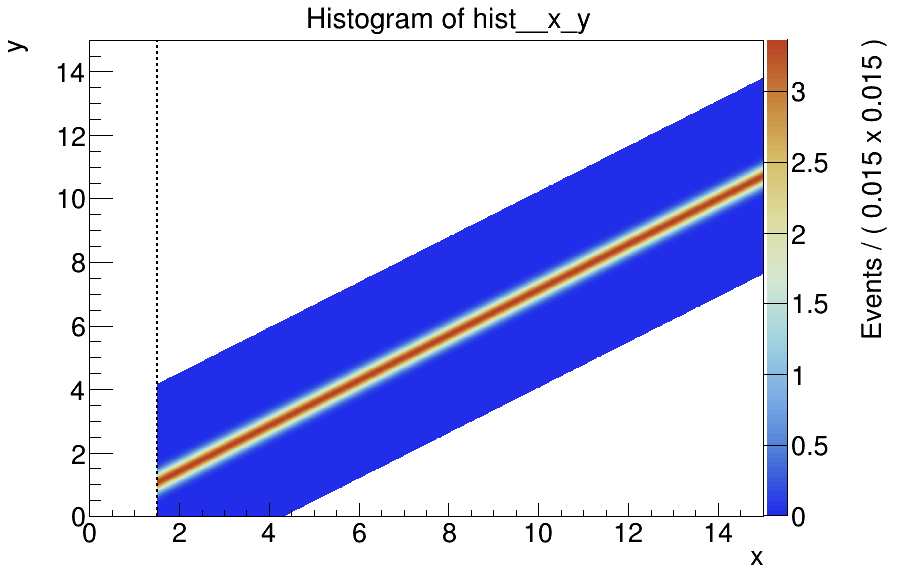
Each of them describes a fit component of a RooAddPdf model in 2D (x, y), by smearing an input TH1F spectrum along a certain line with given resolutions sigma_x, sigma_y. These extended PDFs worked well so far.
But then I wanted to perform a simultaneous fit over several detectors (with different data sets and PDFs), for which I need to add different thresholds in variable x in my model. E.g. detector 1 starts with data from x=0.9, and detector 2 with data from x=1.5. For this I added to my custom PDF a threshold variable, so that the PDF returns 0 if xthreshold. The idea was to have up to 8 different data sets, each only with data for which the x>=threshold for this detector. The corresponding PDFs should then also only start from this threshold.
See the figure showing the PDF with a threshold at 1.5:
With this threshold, the PDFs are now not properly integrated/normalized, at least not for the range and sigma-values I am using (and which I cannot change). Of course a simultaneous fit then fails or gives wrong results.
Here’s an example script to show the problem, the python code and the PDF class can be found in the attachments:
#!/usr/bin/env python
import ROOT
from ROOT import RooFit as RF
ROOT.gSystem.Load("TestPdf_cxx.so")
# Create spectrum and fill it with constant values
spectrum = ROOT.TH1F("spectrum", "spectrum", 100, 0, 30)
for bin in range(1,spectrum.GetNbinsX()+1):
spectrum.SetBinContent(bin, 1.0)
# Create workspace and variables
w = ROOT.RooWorkspace("w")
x = w.factory("x[0,15]")
y = w.factory("y[0,15]")
sigma_x = w.factory("sigma_x[0.1]")
sigma_y = w.factory("sigma_y[0.2]")
threshold = w.factory("threshold[1.5]")
# Create pdf and 2D histogram
pdf = ROOT.TestPdf("pdf", "pdf", x, y, sigma_x, sigma_y, threshold, spectrum)
hist = pdf.createHistogram("hist", x, RF.Binning(1000), RF.YVar(y, RF.Binning(1000)))
# Compare integral of histogram with normalization integral
print "hist integral = ", hist.Integral("WIDTH")
print "pdf integral = ", pdf.expectedEvents( ROOT.RooArgSet(x, y) )
hist.Draw("COLZ")
I already tried using MC integration, but that’s slower and less accurate, which might be a problem for me. Plus I had some problems with this, using more complicated classes:
ROOT.RooAbsReal.defaultIntegratorConfig().method2D().setLabel("RooMCIntegrator")
ROOT.RooAbsReal.defaultIntegratorConfig().getConfigSection("RooMCIntegrator").setRealValue("nIntPerDim",5000)Also I tried increasing the integration precision:
ROOT.RooAbsReal.defaultIntegratorConfig().setEpsAbs(1e-300)
ROOT.RooAbsReal.defaultIntegratorConfig().setEpsRel(1e-300)Is there any other way to improve the integration or my PDF description to make things work? I am using ROOT version 6.05.02 via pyroot.
TestPdf.h (1.47 KB)
TestPdf.cxx (2.27 KB)
test_threshold_pdf.py (1.19 KB)