Dear ROOT and RooFit experts,
I am trying to fit a background shape, which is rapidly falling, and I have a function that I already know it works well.
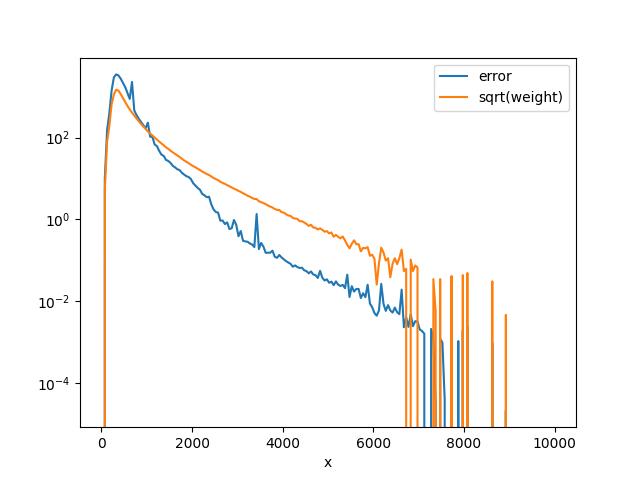
In my analysis I actually need to fit data, but I need to test the function by fitting the simulation (MC). To test the differences that I would see in the case in which I have data, I use the MC and set the error of each bin to sqrt(bin_content). Therefore, I have two MC histograms, both with the same bin content (both in the .root file provided):
- MC whose bin errors are calculated as the sqrt(sumw2)
- MC whose bin errors are calculated as the sqrt(bin_content) (hist_name ending with
sqrtN_error)
I tried fitting both histograms with the RooAbsPdf::fitTo and RooAbsPdf::chi2FitTo functions. The piece of code to fit with the former method is:
# one of the parameters depends on which error we consider
if args.error_type == 'counts':
sumw2_error = False
else:
sumw2_error = True
fit_nll = wksp.pdf(model_name).fitTo(wksp.obj('bkg'), RF.Save(), RF.SumW2Error(sumw2_error), RF.Range('fitrange'))
and the line in which I fit with the chi-square method is:
# none of the parameters depend on the error
fit_chi2 = wksp.pdf(model_name).chi2FitTo(wksp.obj('bkg'), RF.Save(), RF.Range('fitrange'), RF.InitialHesse(True))
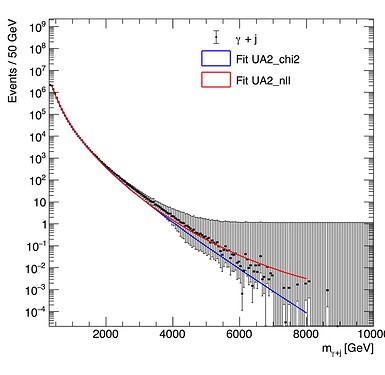
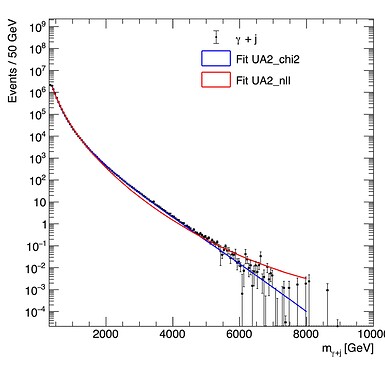
Using the Likelihood method I get the exact same result for both MC histograms (red line), meaning that the different errors are not taken into account (at least that is what I think is happening). In contrast, with the chi-square method I get different results for both the MC histograms (blue line). You can see the fits in the two following figures
I am not sure how can I tell the fitTo method to use the different interpretation of the errors, or to at least get the bin-error information.
Also, using the chi-square method, I noticed that the chi-square value is too high, even in the case of sumw2 errors, in which we can see that the fit is really good.
fit_properties_sumw2_error = {
'model_name' : 'UA2_chi2',
'nvariables' : 4,
'nbins' : 152,
'empty_bins' : 0,
'ndof' : 148,
'chi2' : 1418.446039459683,
'chi2_o_ndof': 9.584094861214075,
'pvalue' : 3.10655364626651e-206
}
fit_properties_counts_error = {
'model_name' : 'UA2_chi2',
'nvariables' : 4,
'nbins' : 152,
'empty_bins' : 0,
'ndof' : 148,
'chi2' : 1651.1243223589468,
'chi2_o_ndof': 11.156245421344234,
'pvalue' : 5.963149078808809e-252
}
To get the chi-square value I am doing:
chi2_var_sumw2_error = model.createChi2(hist,
RF.Range('fitrange'),
RF.DataError(ROOT.RooAbsData.SumW2))
chi2_var_counts_error = model.createChi2(hist,
RF.Range('fitrange'),
RF.DataError(ROOT.RooAbsData.Poisson))
and then I calculate the ndof by hand.
So, to summarize, I have three questions:
- What information should I provide to the
fitTomethod to consider bin-errors appropriately? - Is the
chi2FitTomethod use correctly in both cases of bin-errors? I ask this because the function call doesn’t change, only the errors of the input histogram. - What could be happening to the goodness of fit chi-square and p-values for the case of using the least-squares method, in which I see enormous chi-square values?
Any help on this is greatly appreciated ![]()
Thank you very much for your time and patience
Cheers,
Francisco
input_hists.root (184.0 KB)
bkg_fits.py (4.0 KB)
utils.py (16.0 KB)