Dear experts,
I’m working on a 2D model fit using RooFit. My model includes a signal and two background components (signal, background1, background2) and i want to include a third background component (background3) as a constant contribution.
To test this I generate a dataset from a model without the third background (model_woBG3), afterwards I create a model including the third background (model) and fit it to the dataset.
The intention is, that the third background component stays constant during the fit and the other components are adjusted.
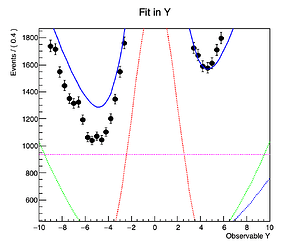
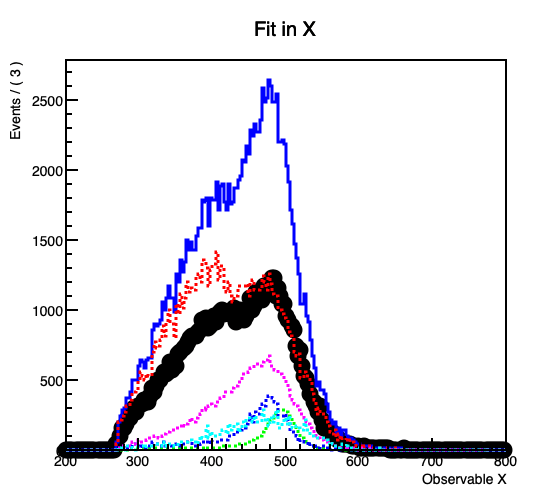
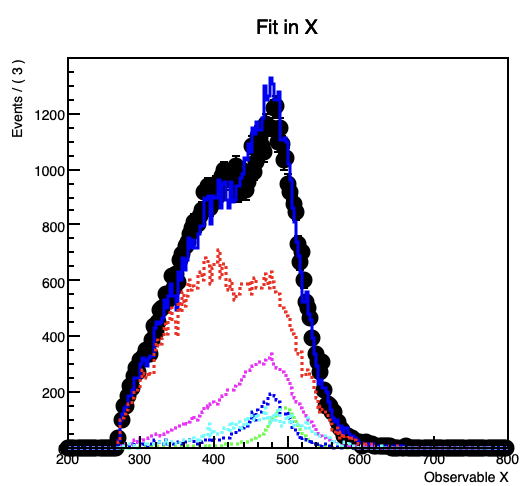
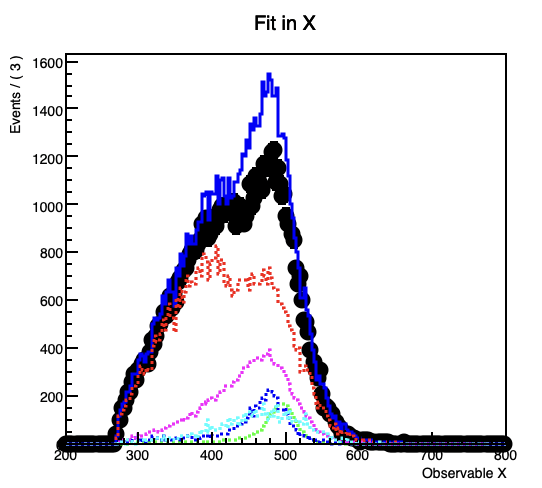
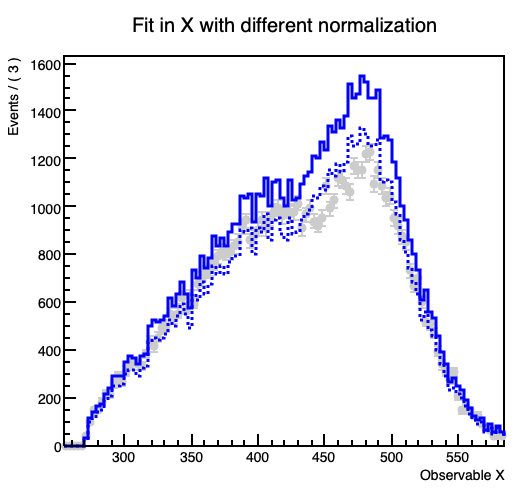
However, when I plot the fitted model and its components (background3) seems to differ slightly from the histogram I get it from, despite declaring nbkg3 as a RooConstVar.
This gets more prominent with increasing the contribution of the the background.
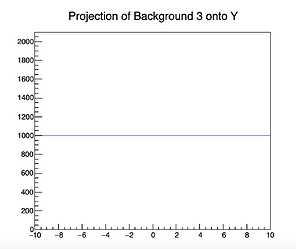
In the code example the projection on the y-Axis from the original background 3 histogram forms a flat line at 1000 entries per bin, while in the fitted model the (background3) component forms a line at about 937.
Here are the relevant lines:
// Signal: gaus in x and y
RooRealVar mean_x("mean_x", "Mean of X", 2, -5, 5);
RooRealVar sigma_x("sigma_x", "Sigma of X", 1, 0.1, 5);
RooGaussian gauss_x("gauss_x", "Signal Gaussian X", x, mean_x, sigma_x);
RooRealVar mean_y("mean_y", "Mean of Y", 0, -5, 5);
RooRealVar sigma_y("sigma_y", "Sigma of Y", 2, 0.1, 5);
RooGaussian gauss_y("gauss_y", "Signal Gaussian Y", y, mean_y, sigma_y);
RooProdPdf signal("signal", "Signal PDF", RooArgList(gauss_x, gauss_y));
// Background 1: exponential in x, 1st-order polynomial in y
RooRealVar bg1_x_tau("bg1_x_tau", "Tau parameter", -0.2, -5., -0.01);
RooExponential bg1_x_expo("bg1_x_expo", "Exponential in X", x, bg1_x_tau);
RooRealVar bg1_y_p0("bg1_y_p0", "Constant term in Y", 0);
RooRealVar bg1_y_p1("bg1_y_p1", "Linear term in Y", 0.1, -1, 1);
RooPolynomial bg1_y_poly("bg1_y_poly", "Polynomial in Y", y, RooArgList(bg1_y_p0, bg1_y_p1));
RooProdPdf background1("background1", "Background 1 PDF", RooArgList(bg1_x_expo, bg1_y_poly));
// Background 2: flat in x, 2nd-order polynomial in y
RooRealVar bg2_x_c0("bg2_x_c0", "Flat in X", 0);
RooPolynomial bg2_x_flat("bg2_x_flat", "Flat Polynomial in X", x, RooArgList(bg2_x_c0));
RooRealVar bg2_y_p1("bg2_y_p1", "Linear term", 0.1, -1, 1);
RooRealVar bg2_y_p2("bg2_y_p2", "Quadratic term", 0.01, -0.5, 0.5);
RooPolynomial bg2_y_poly("bg2_y_poly", "2nd order Polynomial in Y", y, RooArgList(bg2_y_p1, bg2_y_p2));
RooProdPdf background2("background2", "Background 2 PDF", RooArgList(bg2_x_flat, bg2_y_poly));
// Set yields for signal and backgrounds
RooRealVar nsig("nsig", "Number of signal events", 30000, 0, 100000);
RooRealVar nbkg1("nbkg1", "Number of background1 events", 30000, 0, 100000);
RooRealVar nbkg2("nbkg2", "Number of background2 events", 40000, 0, 100000);
// Model without background 3
RooAddPdf model_woBG3("model", "Total Model", RooArgList(signal, background1, background2), RooArgList(nsig, nbkg1, nbkg2));
// Generate a toy dataset
RooDataSet* data = model_woBG3.generate(RooArgSet(x, y), 100000);
// Add background 3 after data set generation (flat in x and y)
TH2* bg3_hist = new TH2F("bg3_hist", "Background 3 Histogram", 50, -10, 10, 50, -10, 10);
for (int i = 0; i < bg3_hist->GetNbinsX(); ++i) {
for (int j = 0; j < bg3_hist->GetNbinsY(); ++j) {
for (int k = 0; k < 20; ++k) {
bg3_hist->Fill(bg3_hist->GetXaxis()->GetBinCenter(i + 1), bg3_hist->GetYaxis()->GetBinCenter(j + 1));
}
}
}
// Create DataHist and PDF for background 3 and set it to be constant
RooDataHist bg3_data("bg3_data", "Background 3 Data", RooArgSet(x, y), bg3_hist);
RooHistPdf background3("background3", "Background 3 PDF", RooArgList(x, y), bg3_data);
RooConstVar nbkg3("nbkg3", "Number of background3 events", bg3_hist->GetEntries());
// RooRealVar nbkg3("nbkg3", "Number of background3 events", bg3_hist->GetEntries());
// nbkg3.setConstant(true);
// Model with background 3
RooAddPdf model("model", "Total Model", RooArgList(signal, background1, background2, background3), RooArgList(nsig, nbkg1, nbkg2, nbkg3));
// Fit model with background 3 to data
RooFitResult* fitResult = model.fitTo(*data);
I also tried using a RooRealVar and setting .setConstant(true), but the issue persisted.
Is there a silly mistake on my side?
I’ve included a working example if anyone would like to try to reproduce the issue.
Any insights, suggestions, or corrections are highly appreciated!
Thanks ![]()
Fit2DConstant.cpp (5.4 KB)