Hi @khsmith, and thanks for bringing this up!
I know the documentation is confusing, but fortunately things are not so complicated in the end. You just have to keep in mind three things:
- When plotting a TH1, the bin content is not divided by bin volume
- When plotting a RooDataHist, the bin content is divided by bin volume to plot the corresponding probability density
- When importing a TH1 to RooDataHist with
RooFit::Import, you can specify that the TH1 content should already be interpreted as a probability density.
So you do everything right  Your histogram represents yields, not probability densities, so indeed using
Your histogram represents yields, not probability densities, so indeed using Import(...,false) is correct 
There problem is that there is a bug in RooFit that the bin width correction is not done correctly when plotting with SumW2 errors! I have opened an issue to fix this, I hope to get this into the next release and maybe also a reduced version of this fix as a backport to existing ROOT releases:
Can you live with Poisson errors for now, or do we need to find a workaround until the fix is merged?
Cheers,
Jonas
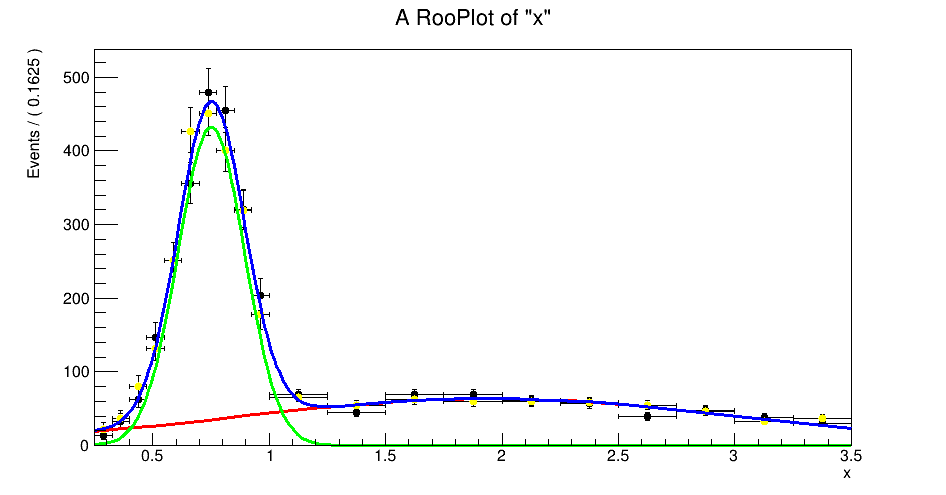
PS: here is example where I do it correctly, with a little cross check: after fitting, I generate a toy dataset from the fit model and plot it together with the original data (yellow vs. black dots). If things are correct, the datasets will be statistically compatible. If you use DataError(RooAbsData::SumW2), this will break until the fix is merged.
std::vector<double> makeBinEdges() {
std::vector<double> binEdges;
for (std::size_t i = 0; i < 10; ++i) {
binEdges.push_back(1 - ((1 - 0.25) / 10) * (10 - i));
}
for (std::size_t i = 0; i < 11; ++i) {
binEdges.push_back(3.5 - ((3.5 - 1.0) / 10) * (10 - i));
}
return binEdges;
}
TH1D* createHisto(std::vector<double> const& binEdges) {
auto histo = new TH1D{"histo",
"histo",
static_cast<int>(binEdges.size() - 1),
binEdges.data()};
for (std::size_t i = 0; i < 1000; ++i) {
histo->Fill(gRandom->Gaus(2.0, 1.0));
}
for (std::size_t i = 0; i < 1000; ++i) {
histo->Fill(gRandom->Gaus(0.75, 0.15));
}
return histo;
}
using namespace RooFit;
// Create the histogram with non-uniform bins
std::vector<double> binEdges = makeBinEdges();
TH1D* histo = createHisto(binEdges);
// Create the model
RooRealVar x{"x", "x", 1.0, binEdges[0], binEdges.back()};
RooRealVar sigMean{
"sigMean", "sigMean", 0.75, binEdges[0], binEdges.back()};
RooRealVar sigSigma{"sigSigma", "sigSigma", 0.15, 0.01, 10.0};
RooRealVar bkgMean{
"bkgMean", "bkgMean", 0.75, binEdges[0], binEdges.back()};
RooRealVar bkgSigma{"bkgSigma", "bkgSigma", 0.15, 0.01, 10.0};
RooRealVar frac{"frac", "frac", 0.5, 0.0, 1.0};
RooGaussian sigGaus{"sigGaus", "sigGaus", x, sigMean, sigSigma};
RooGaussian bkgGaus{"bkgGaus", "bkgGaus", x, bkgMean, bkgSigma};
RooAddPdf model{"model", "model", {sigGaus, bkgGaus}, {frac}};
// Import the histogram to a RooDataHist, where we don't interpred the
// histogram content as densities.
auto* dataHist =
new RooDataHist("dataHist", "dataHist", x, Import(*histo, false));
//Do the fit
RooFitResult* fitRes = model.fitTo(*dataHist, Save(), PrintLevel(-1));
// Generate a toy dataset from the fitted model to compare, veifying that
// the fit to the non-uniform binned dataset was corret.
auto* dataHistToy = model.generate(x, dataHist->sumEntries());
//Plot the full result
TCanvas c{"c", "c"};
RooPlot* frame = x.frame();
dataHist->plotOn(frame, MarkerColor(kBlack));
dataHistToy->plotOn(frame, MarkerColor(kYellow));
model.plotOn(frame, Components("bkgGaus"), LineColor(kRed));
model.plotOn(frame, Components("sigGaus"), LineColor(kGreen));
model.plotOn(frame, LineColor(kBlue));
frame->Draw();
c.SaveAs("plot.png")