Dear RooFit experts,
I am trying to implement a fit with the sum of 3 Gaussians with a 10% resolution uncertainty expressed through a single nuisance parameter (the way the resolution uncertainty is implemented is taken from the internal documentation of H→µµ analysis at CMS).
Below is my Python implementation of the model in RooFit:
# Initialize workspace and the observable variable
w = ROOT.RooWorkspace("w", False)
var = ROOT.RooRealVar("mass","",110,135)
getattr(ROOT.RooWorkspace, 'import')(w, var)
# Mean values of the Gaussians
w.factory("mean1 [125., 120., 130.]")
w.factory("mean2 [125., 120., 130.]")
w.factory("mean3 [125., 120., 130.]")
# Define the value of the uncertainty
w.factory("mu_res_unc [0.1, 0.1, 0.1]")
w.var("mu_res_unc").setConstant(True)
# Gaussian nuisance to be set in the datacard (the value is released after fit)
w.factory("mu_res_beta [0, 0, 0]")
# Width parameters with resolution uncertainty
w.factory("EXPR::width1_times_nuis('width1*(1 + mu_res_unc*mu_res_beta)',{width1[1.0, 0.5, 5.0],mu_res_unc, mu_res_beta})")
w.factory("EXPR::width2_times_nuis('width2*(1 + mu_res_unc*mu_res_beta)',{width2[5.0, 2.0, 10.],mu_res_unc, mu_res_beta})")
w.factory("EXPR::width3_times_nuis('width3*(1 + mu_res_unc*mu_res_beta)',{width3[5.0, 1.0, 10.],mu_res_unc, mu_res_beta})")
# Taking into account shift between mean values of Gaussians
w.factory("expr::deltaM21('mean2-mean1',{mean2, mean1})")
w.factory("expr::deltaM31('mean3-mean1',{mean3, mean1})")
w.factory("EXPR::mean2_final('mean2 + mu_res_unc*mu_res_beta*deltaM21',{mean2, mu_res_unc, mu_res_beta, deltaM21})")
w.factory("EXPR::mean3_final('mean3 + mu_res_unc*mu_res_beta*deltaM31',{mean3, mu_res_unc, mu_res_beta, deltaM31})")
# Gaussians
w.factory("Gaussian::gaus1(mass, mean1, width1_times_nuis)")
w.factory("Gaussian::gaus2(mass, mean2_final, width2_times_nuis)")
w.factory("Gaussian::gaus3(mass, mean3_final, width3_times_nuis)")
gaus1 = w.pdf("gaus1")
gaus2 = w.pdf("gaus2")
gaus3 = w.pdf("gaus3")
# Mixing parameters
w.factory("mix1 [0.5, 0.0, 1.0]")
w.factory("mix2 [0.5, 0.0, 1.0]")
mix1 = w.var("mix1")
mix2 = w.var("mix2")
# Summation
smodel = ROOT.RooAddPdf('smodel', 'smodel', ROOT.RooArgList(gaus1, gaus2, gaus3) , ROOT.RooArgList(mix1, mix2), ROOT.kTRUE)
# Fit the data
smodel.fitTo(data, ROOT.RooFit.Range("full"),ROOT.RooFit.Save(), ROOT.RooFit.Verbose(False))
# Fix the post-fit values of the parameters
for par in ["mean1", "mean2", "mean3", "width1", "width2", "width3", "mix1", "mix2"]:
w.var(par).setConstant(True)
# Release the nuisance parameter
w.var("mu_res_beta").setRange(-5, 5)
w.var("mu_res_beta").setVal(0)
My question is about the summation of the Gaussians:
# Summation
smodel = ROOT.RooAddPdf('smodel', 'smodel', ROOT.RooArgList(gaus1, gaus2, gaus3) , ROOT.RooArgList(mix1, mix2), ROOT.kTRUE)
According to the RooAddPdf documentation (https://root.cern.ch/doc/master/classRooAddPdf.html), the last parameter recursiveFraction defines whether the mixing parameters are interpreted as recursive fraction (and converted into RooRecursiveFraction), or not.
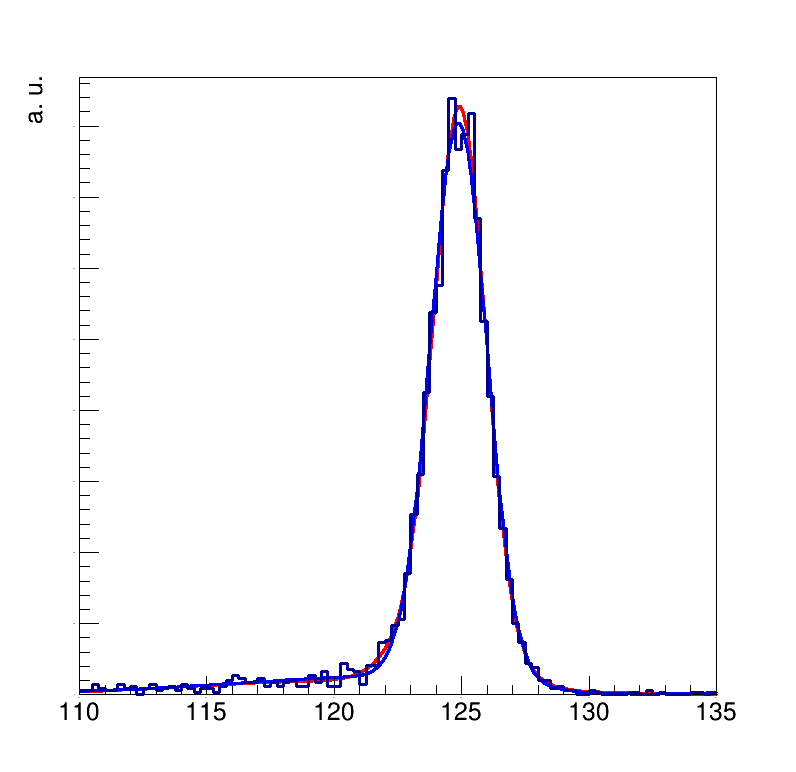
I have tried both setting recursiveFractions=ROOT.kTRUE and recursiveFractions=ROOT.kFALSE and I get slightly different fits: on the attached plot the red line corresponds to kTRUE and the blue line corresponds to kFALSE.
Can you please explain why these two options give different fits, and which option is recommended in this case?
Thanks,
Dmitry