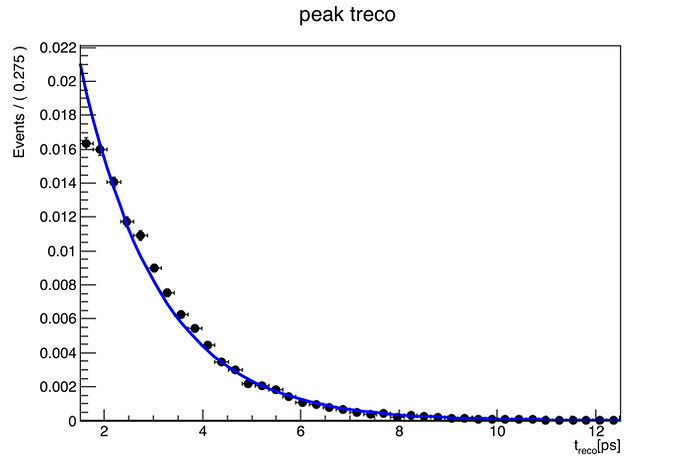
Your macro gives me:
Processing testweight.cc...
RooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
count24171 yield 0.156
entry 18974 weight 0.122458
[#0] WARNING:InputArguments -- RooAbsPdf::fitTo(decay_peak1) WARNING: a likelihood fit is request of what appears to be weighted data.
While the estimated values of the parameters will always be calculated taking the weights into account,
there are multiple ways to estimate the errors on these parameter values. You are advised to make an
explicit choice on the error calculation:
- Either provide SumW2Error(kTRUE), to calculate a sum-of-weights corrected HESSE error matrix
(error will be proportional to the number of events)
- Or provide SumW2Error(kFALSE), to return errors from original HESSE error matrix
(which will be proportional to the sum of the weights)
If you want the errors to reflect the information contained in the provided dataset, choose kTRUE.
If you want the errors to reflect the precision you would be able to obtain with an unweighted dataset
with 'sum-of-weights' events, choose kFALSE.
[#1] INFO:Minization -- RooMinimizer::optimizeConst: activating const optimization
**********
** 1 **SET PRINT 1
**********
**********
** 2 **SET NOGRAD
**********
PARAMETER DEFINITIONS:
NO. NAME VALUE STEP SIZE LIMITS
1 tp1 1.47000e+00 7.35000e-01 0.00000e+00 1.00000e+01
**********
** 3 **SET ERR 0.5
**********
**********
** 4 **SET PRINT 1
**********
**********
** 5 **SET STR 1
**********
NOW USING STRATEGY 1: TRY TO BALANCE SPEED AGAINST RELIABILITY
**********
** 6 **MIGRAD 500 1
**********
FIRST CALL TO USER FUNCTION AT NEW START POINT, WITH IFLAG=4.
START MIGRAD MINIMIZATION. STRATEGY 1. CONVERGENCE WHEN EDM .LT. 1.00e-03
FCN=0.179946 FROM MIGRAD STATUS=INITIATE 4 CALLS 5 TOTAL
EDM= unknown STRATEGY= 1 NO ERROR MATRIX
EXT PARAMETER CURRENT GUESS STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 tp1 1.47000e+00 7.35000e-01 2.14264e-01 -2.62412e-02
ERR DEF= 0.5
MIGRAD MINIMIZATION HAS CONVERGED.
MIGRAD WILL VERIFY CONVERGENCE AND ERROR MATRIX.
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=0.179473 FROM MIGRAD STATUS=CONVERGED 13 CALLS 14 TOTAL
EDM=2.24809e-06 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 tp1 1.59921e+00 5.54951e+00 6.41039e-04 -1.17696e-03
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 1 ERR DEF=0.5
2.180e+01
**********
** 7 **SET ERR 0.5
**********
**********
** 8 **SET PRINT 1
**********
**********
** 9 **HESSE 500
**********
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=0.179473 FROM HESSE STATUS=OK 5 CALLS 19 TOTAL
EDM=2.24805e-06 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER INTERNAL INTERNAL
NO. NAME VALUE ERROR STEP SIZE VALUE
1 tp1 1.59921e+00 5.54951e+00 1.28208e-04 -7.47979e-01
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 1 ERR DEF=0.5
2.180e+01
**********
** 10 **MINOS 500 1
**********
THE POSITIVE MINOS ERROR OF PARAMETER 1, tp1 EXCEEDS ITS LIMIT.
[#0] WARNING:Minization -- RooMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (0.25193) to force MIGRAD to back out of this region. Error log follows
Parameter values: tp1=0
RooDecay::decay_peak1[ t=treco3d tau=tp1 ]
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=1.95641, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=1.95641, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=4.82933, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=4.82933, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=4.32157, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=4.32157, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=1.86453, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=1.86453, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=3.24717, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=3.24717, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=1.73214, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=1.73214, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=1.51113, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=1.51113, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=1.90468, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=1.90468, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=2.37788, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=2.37788, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=8.86509, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=8.86509, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=5.15062, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=5.15062, tau=tp1=0 +/- 5.54951
getLogVal() top-level p.d.f evaluates to zero @ !model=CTM=0, !convVar=treco3d=1.53874, !convSet=(CTM_conv_exp(-@0/@1)_treco3d_tp1_[decay_peak1] = 0/1), t=treco3d=1.53874, tau=tp1=0 +/- 5.54951
... (remaining 2038 messages suppressed)
RooNLLVar::nll_decay_peak1_datap[ paramSet=(tp1) ]
function value is NAN @ paramSet=(tp1 = 0 +/- 5.54951)
FCN=0.179473 FROM MINOS STATUS=PROBLEMS 8 CALLS 27 TOTAL
EDM=2.24805e-06 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER PARABOLIC MINOS ERRORS
NO. NAME VALUE ERROR NEGATIVE POSITIVE
1 tp1 1.59921e+00 5.54951e+00 -1.37246e+00 at limit
ERR DEF= 0.5
[#1] INFO:Minization -- RooMinimizer::optimizeConst: deactivating const optimization
RooFitResult: minimized FCN value: 0.179473, estimated distance to minimum: 2.24805e-06
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0 HESSE=0 MINOS=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
tp1 1.5992e+00 +/- 5.55e+00
[#1] INFO:InputArguments -- RooAbsData::plotOn(datap) INFO: dataset has non-integer weights, auto-selecting SumW2 errors instead of Poisson errors