Dear experts,
I am trying to perform a Wald Test (a.k.a Toy MC test) with a simultaneous PDF model.
The script is in the attached file:
toy_35.cc (6.1 KB)
The PDF models for the simultaneous fit:
conbbsigPDFmom.root (11.8 KB) conbbsigPDFmom_side.root (12.6 KB)
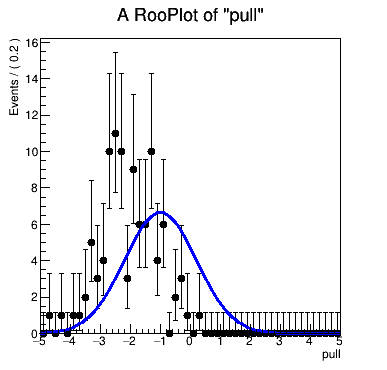
The result is seriously biased:
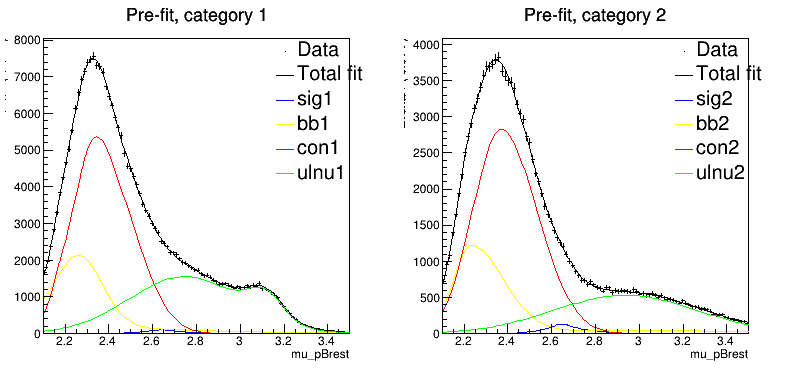
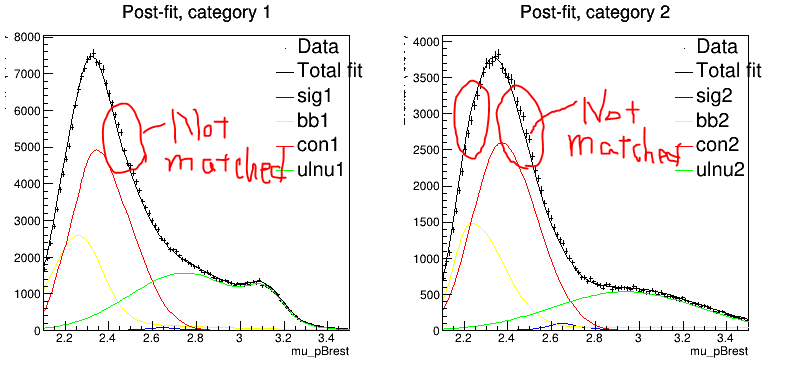
Here comes my question: I tried to plot the pre-fit and post-fit result for one of the generated toy datasets in order to check why the bias occurs. Observing with my naked eyes, I found that the matching between the dataset and the model seems to be even worse after the fit.
Is there anything I do wrong when performing a simultaneous fit?
The code for plotting the pre-fit and post-fit results:
TCanvas *c2 = new TCanvas("c2","c2",800,400);
gStyle->SetErrorX(0.001);
gStyle->SetEndErrorSize(0.);
c2->Divide(2);
c2->cd(1);
RooPlot* frame01 = mu_pBrest.frame(Title("Pre/Post-fit, category 1"));
toyData1->plotOn(frame01,MarkerSize(0.1),Name("mom_side_data"));
Model1.plotOn(frame01,Name("mom_side"),LineColor(1),Slice(cat,"first"),LineWidth(1));
Model1.plotOn(frame01,Name("sigmom_side"),Components(*sig1),Slice(cat,"1"),LineColor(4),LineWidth(1));
Model1.plotOn(frame01,Name("bbmom_side"),Components(*bb1),Slice(cat,"1"),LineColor(5),LineWidth(1));
Model1.plotOn(frame01,Name("conmom_side"),Components(*ulnu1),Slice(cat,"1"),LineColor(2),LineWidth(1));
Model1.plotOn(frame01,Name("ulnumom_side"),Components(*con1),Slice(cat,"1"),LineColor(3),LineWidth(1));
frame01->Draw();
TLegend* leg1 = new TLegend(0.7,0.5,0.95,0.9);
leg1->AddEntry(frame01->findObject("mom_side_data"),"Data","p");
leg1->AddEntry(frame01->findObject("mom_side"),"Total fit","l");
leg1->AddEntry(frame01->findObject("sigmom_side"),"sig1","l");
leg1->AddEntry(frame01->findObject("bbmom_side"),"bb1","l");
leg1->AddEntry(frame01->findObject("conmom_side"),"con1","l");
leg1->AddEntry(frame01->findObject("ulnumom_side"),"ulnu1","l");
leg1->SetTextFont(42);
leg1->SetFillStyle(0);
leg1->SetBorderSize(0);
leg1->Draw("same");
c2->cd(2);
RooPlot* frame02 = mu_pBrest.frame(Title("Pre/Post-fit, category 2"));
toyData2->plotOn(frame02,MarkerSize(0.1),Name("mom_side_data"));
Model2.plotOn(frame02,Name("mom_side"),LineColor(1),Slice(cat,"first"),LineWidth(1));
Model2.plotOn(frame02,Name("sigmom_side"),Components(*sig2),Slice(cat,"2"),LineColor(4),LineWidth(1));
Model2.plotOn(frame02,Name("bbmom_side"),Components(*bb2),Slice(cat,"2"),LineColor(5),LineWidth(1));
Model2.plotOn(frame02,Name("conmom_side"),Components(*ulnu2),Slice(cat,"2"),LineColor(2),LineWidth(1));
Model2.plotOn(frame02,Name("ulnumom_side"),Components(*con2),Slice(cat,"2"),LineColor(3),LineWidth(1));
frame02->Draw();
TLegend* leg2 = new TLegend(0.7,0.5,0.95,0.9);
leg2->AddEntry(frame02->findObject("mom_side_data"),"Data","p");
leg2->AddEntry(frame02->findObject("mom_side"),"Total fit","l");
leg2->AddEntry(frame02->findObject("sigmom_side"),"sig2","l");
leg2->AddEntry(frame02->findObject("bbmom_side"),"bb2","l");
leg2->AddEntry(frame02->findObject("conmom_side"),"con2","l");
leg2->AddEntry(frame02->findObject("ulnumom_side"),"ulnu2","l");
leg2->SetTextFont(42);
leg2->SetFillStyle(0);
leg2->SetBorderSize(0);
leg2->Draw("same");
@StephanH could you please take a look? Thanks!
To me it looks like the model tries to fit something in the tails of the data distribution, where errors are small. This might distort the central region, where you indicated that the fit doesn’t look so good.
How do you define the data errors?
Hello,
I’m not sure what you mean by the “data errors”. Are you asking about the data error in each bin or the error of the size of data?
If you are asking about the definition of data error in each bin, my answer is that this is an unbinned fit, so the bin error is not defined.
If you are talking about the error of the size of data, my answer is that according to the definition of extended UML fit, the distribution of the data size is a Poisson, so the error of the size of data is the Poisson error.
Actually, what really bothers me is the bias of the toy MC. To my knowledge, there can never
be bias in toy MC as long as the data size is large enough. If there is bias in the toy MC, there must be something wrong in the definition of the likelihood function. Thus, when I looked into a single fit and found that in the post-fit result the toy data does not match my PDF, I started to wonder if the log likelihood function is wrong or for some reason it is not minimized at the end of the fit.
This topic was automatically closed 14 days after the last reply. New replies are no longer allowed.