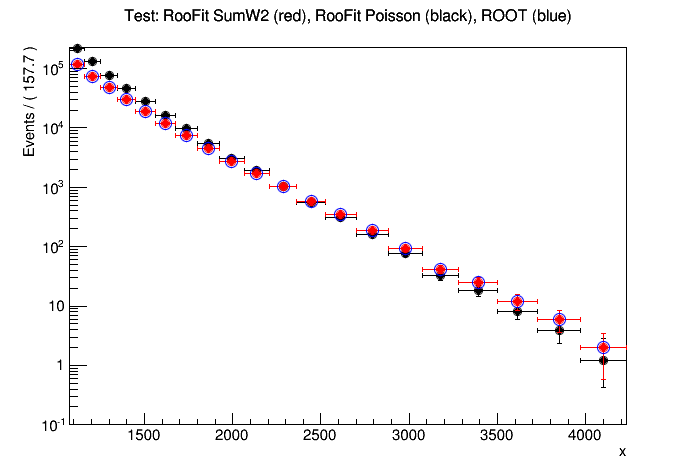
Dear RooFit experts,
I’m having some trouble understanding exactly what happens when asking for plotOn of an imported TH1 with variable bin size when asking for different error configurations. This is not what I’m ultimately trying to do, but I have the feeling it’s connected to the behaviour I’m seeing when trying to perform a chi2 fit to this same histogram.
I’m trying to plot exactly the same histogram, but instead of changing the errors it’s the data points that move when asking for
data.plotOn(xframe,DataError(RooAbsData::Poisson)) instead of
data.plotOn(xframe,DataError(RooAbsData::SumW2)) I don’t see this behaviour when using RooDataSet. The code I am using is below, and the output (test.png) is attached. If needed, I can add the root file to run the example. I’d be grateful if anyone could explain me exactly what is happening here.
Thanks,
Caterina
#include "TFile.h"
#include "TH1D.h"
#include "RooRealVar.h"
#include "RooDataHist.h"
#include "TCanvas.h"
#include "RooPlot.h"
using namespace RooFit;
int main() {
TFile* fin = TFile::Open("dijetMass_data_2012.root","READ");
// extract histogram
TH1D* hist = (TH1D*) fin->Get("mass_0");
// Declare observable x
RooRealVar x("x","x",1076,4100);
//turn the histogram into a RooHist
//do not do density correction so it's comparable to ROOT's own histogram
RooDataHist data(hist->GetName(), hist->GetTitle(), RooArgSet(x), Import(*hist, kFALSE));
// Overlay RooFit...
TCanvas c;
c.Clear();
RooPlot* xframe = x.frame(Title("Test: RooFit SumW2 (red), RooFit Poisson (black), ROOT (blue)")) ;
data.plotOn(xframe,DataError(RooAbsData::Poisson)) ;
data.plotOn(xframe,DataError(RooAbsData::SumW2), MarkerColor(kRed), LineColor(kRed)) ;
xframe->Draw();
gPad->SetLogy();
xframe->SetMinimum(0.1);
//...with Root
hist->SetMarkerColor(kBlue);
hist->SetMarkerStyle(kOpenCircle);
hist->SetMarkerSize(1.5);
hist->Draw("sameP");
c.SaveAs("test.png");
}