Dear ROOTers,
I would like to do a likelihood fit rather than a chisquare fit on the unbinned data with a TGraphAsymmErrors, but I have the impression that this is possible only with histograms.
I explain my problem, maybe I will get also some suggestion about the correct statistical treatment of my problem.
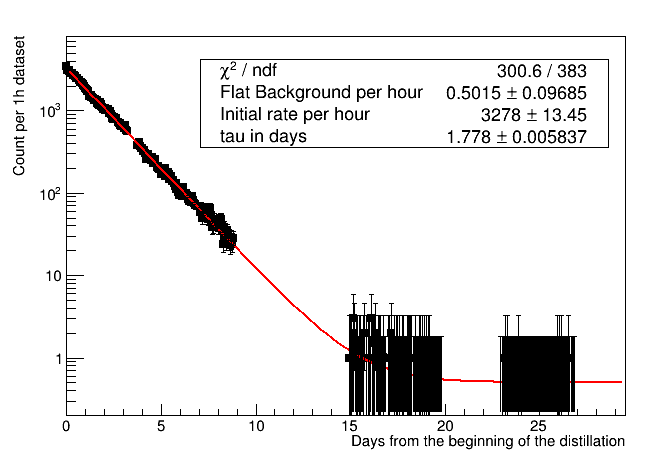
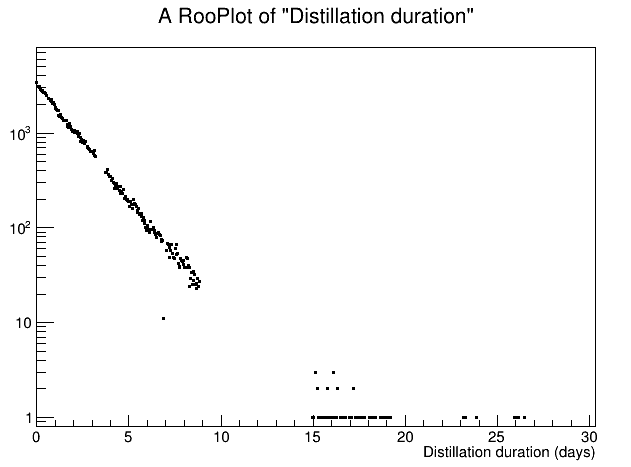
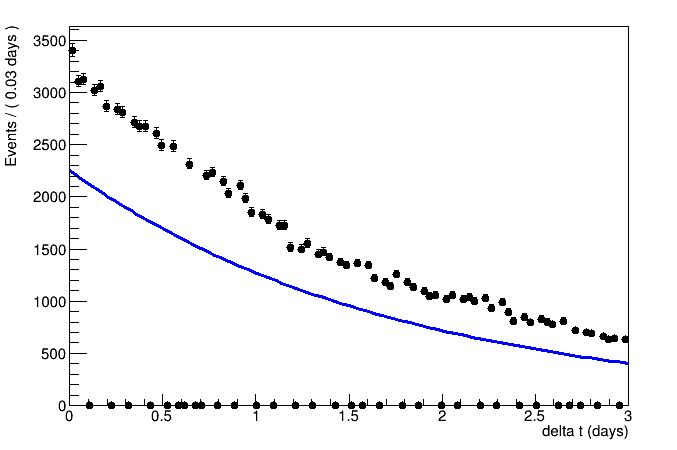
I want to fit the exponential reduction of activity counts over time, very simple (with some expected small flat background). I have datasets of about one hour, but not always contiguous (normally few minutes in between, but there are also few days gaps) and for each of them my selection gives me the counts within this hour. So the rate is the counts divided the “live time” of the dataset, with an error as the sqrt(counts) divided the livetime.
The problem comes from the last “measurement points”, where I get more and more 0 counts.
I do not see an option for max likelihood fit in TGraphAsymmErrors and chisquare fits cannot take into accounts zeros. Moreover already when I have few counts per datasets I would like to use the more accurate asymmetric “poisson” error.
So far my workaround is to group together datasets in days, but now I have the same problem in the last few days, and this is the period where I start to see a change of slope and I need to figure out if it is really a change of slope or a background larger than expected.
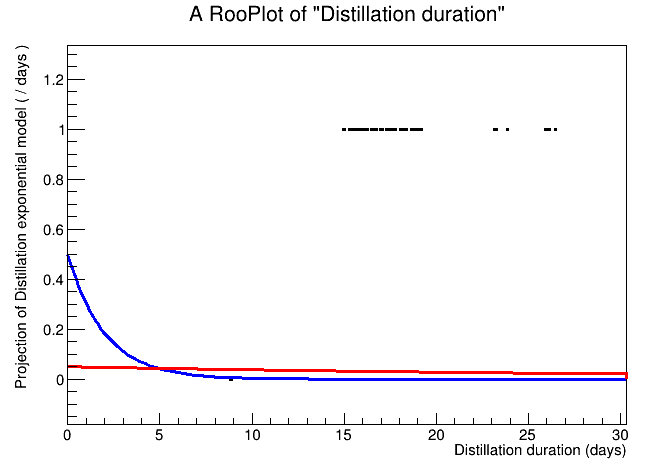
One solution could be to make a TH1D with variable bin size to place the measurement points at the center of each bin, but 1) I am not sure how this variable bin size enters into the likelihood 2) the few days gaps are either a wrong zero data points or they move the center of the adjacent bin if I decide to merge them to the next bin.
If there are no options with TH1 or TGraphAsymmError, I have quickly read that RooFit performs unbinned max likelihood fit and I wonder if this is the way to go (but I have to learn the tool…)
Any suggestion is very welcome,
Matteo
ROOT Version: 6.14/02 or master, but I doubt that this matters in this case
Platform: Debian 9
Compiler: gcc 6.3