Dear All,
My code is working well but now I want to do some modification that seems to be trivial but I have some trouble to do it.
In fact I am using a file txt named lowmass_a0a0 to plot in a empty graph , now in the same graph I want to do another plot from another file txt named highmass_a0a0 for instance. I guess a loop that can do the same of what the code is doing now should be enough but it has just to do it with the highmass_a0a0 file. I attached the code.
Would apreciate if soemeone could help.
CheersBRHaalimitSetHigh_low_mass.C (6.3 KB)
Can you post a macro we can run ?
Thanks
I think we just need to input files (although I would need these special headers too).
Hi Everyone,
Thanks for replying, yes at the end I just input the files and create a vectors about the number of points since the files have not the same number of lines.
Then I replace this for loop for(Int_t i=0;i<1;i++){…} by this one for(Int_t i=0;i<2;i++){…} but I put the canva and the empty graph before this for loop so it will not be repeated. And it works.
Thanks a lot.
Cheers
Hi Everyone,
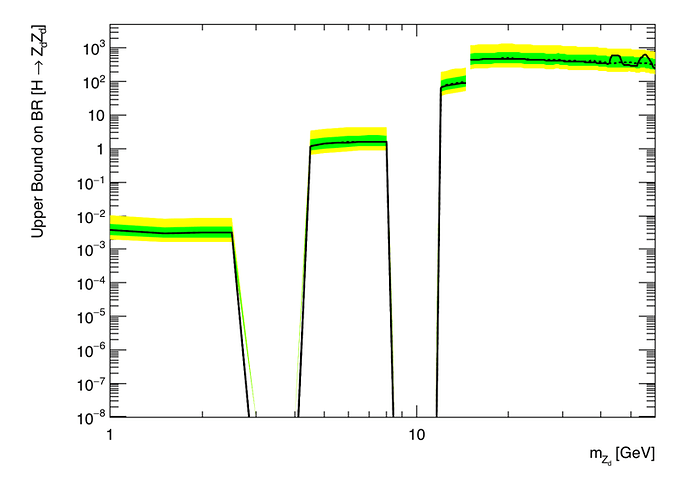
In fact I realized that my problem is only partially resolved. In fact I would like to do the plot using log scale on Y and X axis. I am having error if I use log scale on Y axis that I can understand because there are some zero in Y axis for instance for 3 <= mZd <= 4 and 8.5 <= mZd <= 11.5 the values on Y axis are zero. So I would like hatch these two regions and make a vertical line between low and high mass (low mass goes from zero to 15 and high mass from 15 to 60).
#include "Math/Interpolator.h"
#include <iostream>
#include "AtlasStyle.C"
void BRHaalimitSetHigh_low_mass() {
TString files_lm[]={"./LowMass_a0a0.txt","HighMass_a0a0.txt"};
// TString files_hm[]={"./HighMass_a0a0.txt"};
TString files[]={"./type-1.dat"};
// TString s[]={"3"};
TString canv[]={"can3"};
gSystem->Load("AtlasStyle.C");
SetAtlasStyle();
// const unsigned int Npoints_lm = 28;
std::vector<unsigned int> Npoints {28,91};
// Npoints[0]= 28;
// Npoints[1]= 91;
// const unsigned int Npoints_hm = 91;
const unsigned int Npoints_fix = 673;
const unsigned int Npoints_max = 1000;
Double_t mzd[Npoints_max];
Double_t Brall[Npoints_max];
Double_t ggFSigma[Npoints_max];
Double_t fac[Npoints_max];
double mass[Npoints_max];
double obs[Npoints_max];
double Exp[Npoints_max];
double onesiglow[Npoints_max];
double onesighigh[Npoints_max];
double twosiglow[Npoints_max];
double twosighigh[Npoints_max];
TCanvas *can = new TCanvas(canv[0], canv[0],66,52,800,600);
can->Range(0,0,1,1);
can->SetFillColor(0);
can->SetBorderMode(0);
can->SetBorderSize(2);
can->SetFrameBorderMode(0);
can->SetLogx();
can->SetLogy();
//Plot the observed limits
TGraphErrors *gre = new TGraphErrors(Npoints_max);
gre->SetName("CLs_observed");
gre->SetTitle("Observed CLs");
gre->SetFillColor(1);
gre->SetLineColor(1);
gre->SetLineWidth(2);
gre->SetMarkerStyle(20);
// cout<<Exp[0]*(ggFSigma[0])<<endl;
TH1F *Graph_CLs_observed1 = new TH1F("Graph_CLs_observed1","",100,1,60);
Graph_CLs_observed1->SetMinimum(1.0e-8);
Graph_CLs_observed1->SetMaximum(5000);
Graph_CLs_observed1->SetDirectory(0);
Graph_CLs_observed1->SetStats(0);
TGaxis *yaxis = (TGaxis*)Graph_CLs_observed1->GetYaxis();
yaxis->SetMaxDigits(2);
Int_t ci; // for color index setting
Int_t cii;
ci = TColor::GetColor("#000099");
Graph_CLs_observed1->SetLineColor(ci);
Graph_CLs_observed1->GetXaxis()->SetTitle("m_{Z_{d}} [GeV]");
Graph_CLs_observed1->GetXaxis()->SetLabelFont(42);
Graph_CLs_observed1->GetXaxis()->SetLabelSize(0.035);
Graph_CLs_observed1->GetXaxis()->SetTitleSize(0.035);
Graph_CLs_observed1->GetXaxis()->SetTitleFont(42);
Graph_CLs_observed1->GetYaxis()->SetTitle("Upper Bound on BR [H #rightarrow Z_{d}Z_{d}]");
Graph_CLs_observed1->GetYaxis()->SetLabelFont(42);
Graph_CLs_observed1->GetYaxis()->SetLabelSize(0.035);
Graph_CLs_observed1->GetYaxis()->SetTitleSize(0.035);
Graph_CLs_observed1->GetYaxis()->SetTitleFont(42);
Graph_CLs_observed1->GetYaxis()->SetTitleOffset(1.85);
Graph_CLs_observed1->GetZaxis()->SetLabelFont(42);
Graph_CLs_observed1->GetZaxis()->SetLabelSize(0.035);
Graph_CLs_observed1->GetZaxis()->SetTitleSize(0.035);
Graph_CLs_observed1->GetZaxis()->SetTitleFont(42);
gre->SetHistogram(Graph_CLs_observed1);
gre->Draw("al");
//Plot for the expected limits and the pm 1 and 2 sigma fluctuations
TMultiGraph *multigraph = new TMultiGraph();
multigraph->SetName("Plot_expected");
multigraph->SetTitle("Expected Plot");
for(Int_t i=0;i<2;i++){
TString tmps;
ifstream fin_lm(files_lm[i]);
ifstream fin(files[0]);
for (Int_t k=0; k<Npoints_fix; k++)
{
fin>>tmps;mzd[k]=tmps.IsFloat()?tmps.Atof():0;fin>>tmps; Brall[k]=tmps.IsFloat()?tmps.Atof():0;
}
// cout<<Brall[672]<<endl;
ROOT::Math::Interpolator inter(673, ROOT::Math::Interpolation::kCSPLINE);
inter.SetData(673, mzd, Brall);
double invCrossSection = (1.0 / 48.59)*0.001;
// fin>>tmps;fin>>tmps;fin>>tmps;fin>>tmps;fin>>tmps;fin>>tmps;fin>>tmps;//read first line.
for (Int_t j=0; j<Npoints[i]; j++)
{
fin_lm>>tmps;mass[j]=tmps.IsFloat()?tmps.Atof():0;fin_lm>>tmps; obs[j]=tmps.IsFloat()?tmps.Atof():0; fin_lm>>tmps; Exp[j]=tmps.IsFloat()?tmps.Atof():0; fin_lm>>tmps; onesiglow[j]=tmps.IsFloat()?tmps.Atof():0; fin_lm>>tmps; onesighigh[j]=tmps.IsFloat()?tmps.Atof():0; fin_lm>>tmps; twosiglow[j]=tmps.IsFloat()?tmps.Atof():0; fin_lm>>tmps; twosighigh[j]=tmps.IsFloat()?tmps.Atof():0;
double interBr = inter.Eval( mass[j] );
fac[j] = invCrossSection/ (interBr*interBr);
ggFSigma[j] = fac[j];
// cout<<ggFSigma[j]<<endl;
}
TGraphAsymmErrors *grae = new TGraphAsymmErrors(Npoints[i]);
grae->SetName("sig2");
grae->SetTitle("Expected CLs #pm 2 #sigma");
ci = TColor::GetColor("#ffff00");
grae->SetFillColor(ci);
for ( Int_t j=0; j<Npoints[i]; j++) {
// if (mass[j] != 0){
grae->SetPoint(j,mass[j],Exp[j]*(ggFSigma[j]));
grae->SetPointError(j,0,0,Exp[j]*(ggFSigma[j])-twosiglow[j]*(ggFSigma[j]),twosighigh[j]*(ggFSigma[j])-Exp[j]*(ggFSigma[j]));
// cout<<Exp[j] <<endl;}
}
multigraph->Add(grae,"3");
grae = new TGraphAsymmErrors(22);
grae->SetName("sig1");
grae->SetTitle("Expected CLs #pm 1 #sigma");
ci = TColor::GetColor("#00ff00");
grae->SetFillColor(ci);
for ( Int_t j=0; j<Npoints[i]; j++ ) {
grae->SetPoint(j,mass[j],Exp[j]*(ggFSigma[j]));
grae->SetPointError(j,0,0,Exp[j]*(ggFSigma[j])-onesiglow[j]*(ggFSigma[j]),onesighigh[j]*(ggFSigma[j])-Exp[j]*(ggFSigma[j]));
}
multigraph->Add(grae,"3");
TGraph *graph = new TGraph(Npoints[i]);
graph->SetName("");
graph->SetTitle("Expected CLs - Median");
graph->SetFillColor(1);
graph->SetLineColor(1);
graph->SetLineStyle(2);
graph->SetLineWidth(2);
for (Int_t j=0; j<Npoints[i]; j++) {
graph->SetPoint(j,mass[j],Exp[j]*(ggFSigma[j]));
}
multigraph->Add(graph,"L");
TGraph *graphN = new TGraph(Npoints[i]);
graphN->SetName("SM_crosssections");
graphN->SetTitle("Total SM Cross-Sections");
graphN->SetFillColor(1);
graphN->SetLineColor(2);
graphN->SetLineStyle(2);
graphN->SetLineWidth(2);
for ( Int_t j=0; j<Npoints[i]; j++) {
graphN->SetPoint(j,mass[j],(ggFSigma[j]));
}
// multigraph->Add(graphN,"L");
//multigraph->Draw("");
gre = new TGraphErrors(Npoints[i]);
gre->SetName("CLs_observed");
gre->SetTitle("Observed CLs");
gre->SetFillColor(1);
gre->SetLineColor(1);
gre->SetLineWidth(2);
gre->SetMarkerStyle(20);
for ( Int_t j=0; j<Npoints[i]; j++) {
gre->SetPoint(j,mass[j],obs[j]*(ggFSigma[j]));
gre->SetPointError(j,0,0);
// cout<<obs[j]*(ggFSigma[j])<<endl;
}
multigraph->Add(gre,"l");
// gre->Draw("l");
multigraph->Draw("");
}
TLegend *leg = new TLegend(0.7,0.7,0.9,0.9,NULL,"brNDC");
leg->SetBorderSize(1);
leg->SetLineColor(1);
leg->SetLineStyle(1);
leg->SetLineWidth(1);
leg->SetFillColor(19);
leg->SetFillStyle(1001);
TLegendEntry * entry=leg->AddEntry("CLs_observed","Observed","L");
entry->SetLineColor(1);
entry->SetLineStyle(1);
entry->SetLineWidth(2);
entry->SetMarkerColor(1);
entry->SetMarkerStyle(20);
entry->SetMarkerSize(1);
entry=leg->AddEntry("Plot_expected","Expected","l");
entry->SetLineColor(1);
entry->SetLineStyle(2);
entry->SetLineWidth(2);
entry->SetMarkerColor(1);
entry->SetMarkerStyle(21);
entry->SetMarkerSize(1);
entry=leg->AddEntry("sig1","#pm 1 #sigma","F");
ci = TColor::GetColor("#00ff00");
entry->SetFillColor(ci);
entry->SetFillStyle(1001);
entry->SetLineColor(1);
entry->SetLineStyle(1);
entry->SetLineWidth(1);
entry->SetMarkerColor(3);
entry->SetMarkerStyle(21);
entry->SetMarkerSize(1);
entry=leg->AddEntry("sig2","#pm 2 #sigma","F");
ci = TColor::GetColor("#ffff00");
entry->SetFillColor(2);
entry->SetFillStyle(1001);
entry->SetLineColor(1);
entry->SetLineStyle(1);
entry->SetLineWidth(1);
entry->SetMarkerColor(1);
entry->SetMarkerStyle(21);
entry->SetMarkerSize(1);
// leg->Draw();
TLatex *tex = new TLatex();
/* tex->SetTextFont(72);
tex->DrawLatex(17,3.0e-4,"ATLAS");
tex->SetTextFont(42);
tex->DrawLatex(24.5,3.0e-4,"Internal");
tex->DrawLatex(17,3.0e-4,"#sqrt{s} = 13 TeV, 36.5 fb^{-1}");
tex->DrawLatex(17,2.8e-4,"95% CL");
// tex->DrawLatex(17,2.6e-4,"Final State: 4e+2e2#mu+4#mu");
tex->DrawLatex(17,2.6e-4,"Final State: 4#mu");*/
}
For instance here is the plot I got (even with the error caused by the zero on the Y axis).
Here is my code for instance:LowMass_a0a0.txt (1.5 KB)
HighMass_a0a0.txt (4.9 KB)
Thanks in advance.
Cheers
Your macro gives the following output and an empty plot:
Processing BRHaalimitSetHigh_low_mass.C...
Error in <GSLError>: Error 4 in interp.c at 83 : x values must be strictly increasing
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: Suppressing additional warnings
Error in <GSLError>: Error 4 in interp.c at 83 : x values must be strictly increasing
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: Suppressing additional warnings
Hi Couet,
Thanks for replying.
It’s a bit strange but it gives the plot I attached, may be we don’t have the same root version. I am using ROOT 6.08/06, or maybe you need these AtlasStyle files, which I am going to attach here. AtlasStyle.h (388 Bytes)
AtlasStyle.C (2.5 KB)
Cheers
I get the same thing with these two files. I tried root master and 6.08/07. It seems to be a Math issue. May be @moneta will have an idea.
Hi Couet,
I think you are right it’s a math issue, I remember I was having the same issue but I did this to resolve it:
cmake -Dmathmore=ON /opt/root/root-6.08.06 -DCMAKE_INSTALL_PREFIX=/opt/root/root-6.08.06_install/
where /opt/root/root-6.08.06 is the path where I build and install root, and /opt/root/root-6.08.06_install/ is the directory where I install root.
then I did this: make -j2
and this: sudo make install
It will effect only Math stuff in root.
Cheers
…Did you try it?
Cheers
yes (I had -D=All anyway) … And I get the same:
$ root
-------------------------------------------------------------------------
| Welcome to ROOT 6.08/07 http://root.cern.ch |
| (c) 1995-2017, The ROOT Team |
| Built for macosx64 |
| From heads/v6-08-00-patches@v6-08-06-68-gff9a7c0, May 22 2017, 13:00:39 |
| Try '.help', '.demo', '.license', '.credits', '.quit'/'.q' |
-------------------------------------------------------------------------
root [0] .x BRHaalimitSetHigh_low_mass.C
Applying ATLAS style settings...
Error in <GSLError>: Error 4 in interp.c at 83 : x values must be strictly increasing
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Error in <GSLError>: Error 4 in interp.c at 83 : x values must be strictly increasing
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
Warning in <ROOT::Math::GSLInterpolator::Eval>: input domain error
root [1]
As I said I let @moneta answer you.
Ok cool, let’s wait for him, I keep trying to find a solution meanwhile.
Thanks
Cheers
This topic was automatically closed 14 days after the last reply. New replies are no longer allowed.