Hi @moneta, @jonas I think I’m being an idiot and missing something, can you help out?
I’d like to do a simple likelihood ratio test to say, “is the data closer to H0 or H1” I have the HistFactory model, H0 == [norm0 = 1, norm1 = 0] and H1 == [norm0 = 0, norm1 = 1] (weird that I have to set parameters for this but ok maybes it’s relevant)
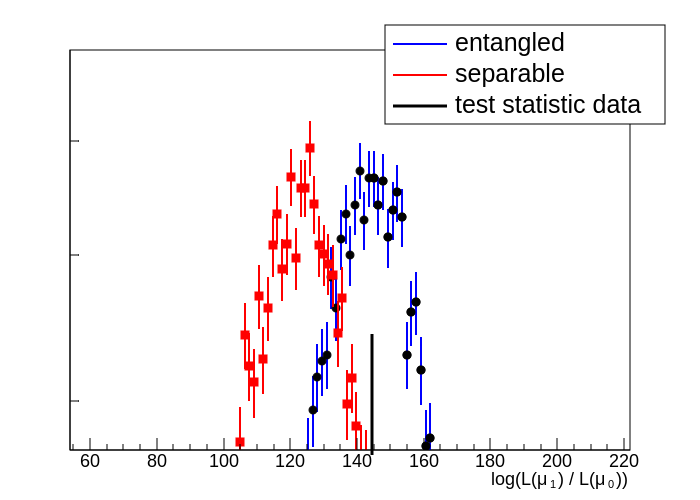
I can get the regular hypo test plot to work and it looks like this:
Which is pretty good and of course produced with the standard RooStats tutorial code:
slrts = ROOT.RooStats.SimpleLikelihoodRatioTestStat(sModel.GetPdf(), eModel.GetPdf())
nullParams = sModel.GetSnapshot()
nullParams.addOwned(sModel.GetNuisanceParameters())
slrts.SetNullParameters(nullParams)
altParams = eModel.GetSnapshot()
altParams.addOwned(eModel.GetNuisanceParameters())
slrts.SetAltParameters(altParams)
hypoCalc = ROOT.RooStats.FrequentistCalculator(data, eModel, sModel)
hypoCalc.SetToys(10000,10000)
hypoCalc.StoreFitInfo(True)
sampler = hypoCalc.GetTestStatSampler()
sampler.SetGenerateBinned(True)
sampler.SetTestStatistic(slrts)
htr = hypoCalc.GetHypoTest()
plot = ROOT.RooStats.HypoTestPlot(htr, 100)
c = ROOT.TCanvas()
plot.SetLogYaxis(True)
plot.Draw()
c.Draw()
but I would like to extract out the likelihood ratio function itself to extend the plot and calculate the values directly. That should be simple enough right?
If I do a simple createNLL() on either of the PDFs it seems to perform a fit…
def assign_snapshot(pdf, data, snapshot):
“”“Assign snapshot values to the parameters of `pdf` relevant for `data`.”“”
params = pdf.getParameters(data)
for p in params:
p.Print()
val = snapshot.find(p)
if val:
p.setVal(val.getVal())
def binned_nll_at_snapshot(pdf, data, snapshot):
assign_snapshot(pdf, data, snapshot)
nll = pdf.createNLL(
data,
ROOT.RooFit.Extended(True),
ROOT.RooFit.Offset(True)
)
val = nll.getVal()
del nll
return val
pdf0 = sModel.GetPdf()
pdf1 = eModel.GetPdf()
data = ws.data(“obs_data”)
nll0 = binned_nll_at_snapshot(pdf0, data, sModel.GetSnapshot())
nll1 = binned_nll_at_snapshot(pdf1, data, sModel.GetSnapshot())
firstly they give near identical values (always with a ratio around 1.00 not the 142 shown in the plot), but more worryingly I get the following:
[#1] INFO:Minimization – Including the following constraint terms in minimization: (lumiConstraint,alpha_Constraint,gamma_stat_cCGLMP_channel_bin_0_constraint,gamma_stat_cCGLMP_channel_bin_1_constraint,gamma_stat_cCGLMP_channel_bin_2_constraint,gamma_stat_cCGLMP_channel_bin_3_constraint,gamma_stat_cCGLMP_channel_bin_4_constraint,gamma_stat_cCGLMP_channel_bin_5_constraint,gamma_stat_cCGLMP_channel_bin_6_constraint,gamma_stat_cCGLMP_channel_bin_7_constraint,gamma_stat_cCGLMP_channel_bin_8_constraint,gamma_stat_cCGLMP_channel_bin_9_constraint,gamma_stat_cCGLMP_channel_bin_10_constraint,gamma_stat_cCGLMP_channel_bin_11_constraint,gamma_stat_cCGLMP_channel_bin_12_constraint,gamma_stat_cCGLMP_channel_bin_13_constraint,gamma_stat_cCGLMP_channel_bin_14_constraint,gamma_stat_cCGLMP_channel_bin_15_constraint,gamma_stat_cCGLMP_channel_bin_16_constraint,gamma_stat_cCGLMP_channel_bin_17_constraint,gamma_stat_cCGLMP_channel_bin_18_constraint,gamma_stat_cCGLMP_channel_bin_19_constraint,gamma_stat_cCGLMP_channel_bin_20_constraint,gamma_stat_cCGLMP_channel_bin_21_constraint,gamma_stat_cCGLMP_channel_bin_22_constraint,gamma_stat_cCGLMP_channel_bin_23_constraint,gamma_stat_cCGLMP_channel_bin_24_constraint,gamma_stat_cCGLMP_channel_bin_25_constraint,gamma_stat_cCGLMP_channel_bin_26_constraint,gamma_stat_cCGLMP_channel_bin_27_constraint,gamma_stat_cCGLMP_channel_bin_28_constraint,gamma_stat_cCGLMP_channel_bin_29_constraint,gamma_stat_cCGLMP_channel_bin_30_constraint,gamma_stat_cCGLMP_channel_bin_31_constraint,gamma_stat_cCGLMP_channel_bin_32_constraint,gamma_stat_cCGLMP_channel_bin_33_constraint,gamma_stat_cCGLMP_channel_bin_34_constraint,gamma_stat_cCGLMP_channel_bin_35_constraint,gamma_stat_cCGLMP_channel_bin_36_constraint,gamma_stat_cCGLMP_channel_bin_37_constraint,gamma_stat_cCGLMP_channel_bin_38_constraint)
[#1] INFO:Minimization – The global observables are not defined , normalize constraints with respect to the parameters (Lumi,alpha_,f,f_sep,gamma_stat_cCGLMP_channel_bin_0,gamma_stat_cCGLMP_channel_bin_1,gamma_stat_cCGLMP_channel_bin_10,gamma_stat_cCGLMP_channel_bin_11,gamma_stat_cCGLMP_channel_bin_12,gamma_stat_cCGLMP_channel_bin_13,gamma_stat_cCGLMP_channel_bin_14,gamma_stat_cCGLMP_channel_bin_15,gamma_stat_cCGLMP_channel_bin_16,gamma_stat_cCGLMP_channel_bin_17,gamma_stat_cCGLMP_channel_bin_18,gamma_stat_cCGLMP_channel_bin_19,gamma_stat_cCGLMP_channel_bin_2,gamma_stat_cCGLMP_channel_bin_20,gamma_stat_cCGLMP_channel_bin_21,gamma_stat_cCGLMP_channel_bin_22,gamma_stat_cCGLMP_channel_bin_23,gamma_stat_cCGLMP_channel_bin_24,gamma_stat_cCGLMP_channel_bin_25,gamma_stat_cCGLMP_channel_bin_26,gamma_stat_cCGLMP_channel_bin_27,gamma_stat_cCGLMP_channel_bin_28,gamma_stat_cCGLMP_channel_bin_29,gamma_stat_cCGLMP_channel_bin_3,gamma_stat_cCGLMP_channel_bin_30,gamma_stat_cCGLMP_channel_bin_31,gamma_stat_cCGLMP_channel_bin_32,gamma_stat_cCGLMP_channel_bin_33,gamma_stat_cCGLMP_channel_bin_34,gamma_stat_cCGLMP_channel_bin_35,gamma_stat_cCGLMP_channel_bin_36,gamma_stat_cCGLMP_channel_bin_37,gamma_stat_cCGLMP_channel_bin_38,gamma_stat_cCGLMP_channel_bin_4,gamma_stat_cCGLMP_channel_bin_5,gamma_stat_cCGLMP_channel_bin_6,gamma_stat_cCGLMP_channel_bin_7,gamma_stat_cCGLMP_channel_bin_8,gamma_stat_cCGLMP_channel_bin_9)
[#1] INFO:Fitting – RooAbsPdf::fitTo(cCGLMP_channel_model) fixing normalization set for coefficient determination to observables in data
my ‘data’ here is just the Asimov data according to H1 (obviously) so if it fits both PDFs (which are parametric because otherwise RooStats complains) to give the norm0=0 and norm1=1 coefficients that produced the data and making the whole thing pointless.
Am I missing a flag or something here? I feel like this isn’t the kind of thing someone like me should be asking…