Dear experts,
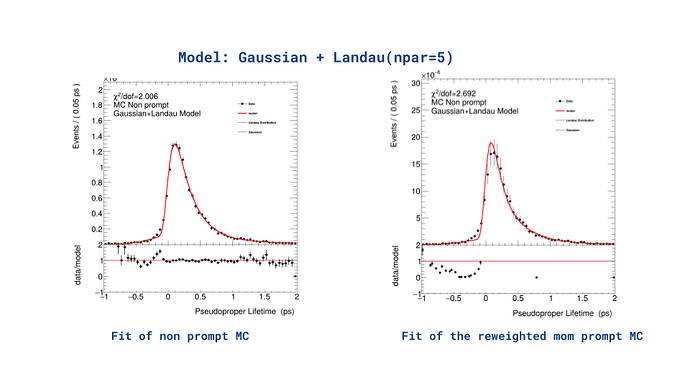
I am encountering an issue with the residual plot when fitting a model to reweighted data. Initially, I worked with MC data, applied a model, and the fit converged successfully with a reasonable residual plot.
Later, I reweighted the MC data based on a measurement. As part of this process, I obtained bin contents and errors, which I used to create a new reweighted histogram. I then reused the same fitting code, changing only the histogram input. The fit still converges well; however, the residual plot (data/model) now shows strange behavior—some points appear abnormal, and others are missing entirely especially from 0.8 to 1.95.
Interestingly, when I switch back to the original MC histogram, everything works as expected. This leads me to suspect that the issue may lie in how I’m filling the reweighted histogram, since the fitting code itself seems to work perfectly for the original MC histogram.
Any insight or suggestions would be greatly appreciated.
Thank you in advance
Code I’m using to create the reweighted histogram
nphistweight.py (2.4 KB)
Code used for the fit pyroot rofit
npfitw.py (6.1 KB)