Hi Stephan,
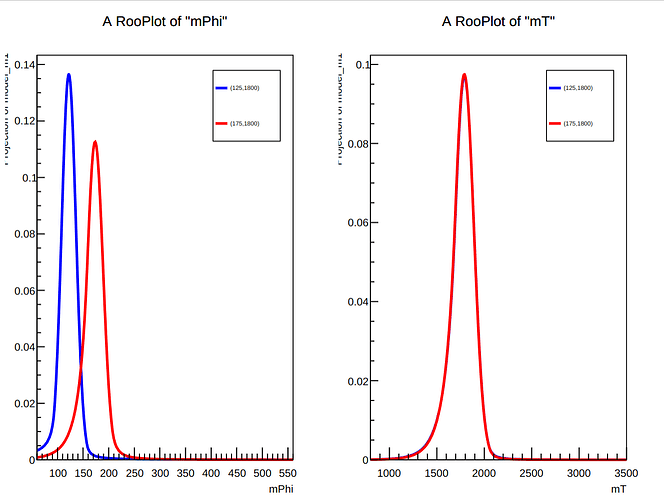
Thanks very much for the quick reply. I think the error was somewhere in the creation of the RooDataHists, but I’m not sure where exactly. I worked around it by fitting the two existing signals (125, 1800) and (175, 1800) with a product of RooCrystalBall functions along the two axes and saving the resulting RooProdPdf to a RooWorkspace in a ROOT file. This works well, as evidenced by the fit result plot of the PDFs:
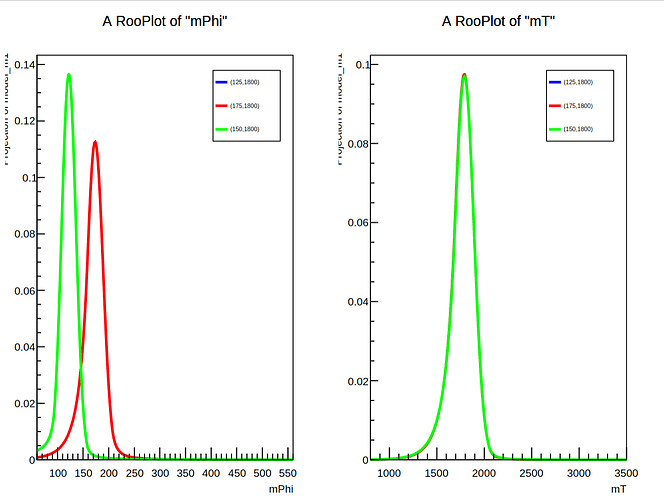
However, when I attempt to perform the interpolation for, e.g, a signal at (150, 1800), it seems only to copy the signal at (125, 1800) instead of interpolating between mPhi=125 and mPhi=175.
There are some messages from RooFit printed to the terminal (the error messages I believe can be ignored, as per here):
------------------------------------------------------------------------------------------
Interpolating signal of mPhi = 150, between existing signals at mPhi = [125,175]
------------------------------------------------------------------------------------------
[#1] INFO:NumericIntegration -- RooRealIntegral::init(model_MOMENT_1_mPhi_product_Int[mPhi,mT]_Norm[mPhi,mT]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(mPhi,mT)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(model_MOMENT_1_mT_product_Int[mPhi,mT]_Norm[mPhi,mT]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(mT,mPhi)
[#0] ERROR:InputArguments -- RooArgSet::checkForDup: ERROR argument with name model_MOMENT_2C_mPhi is already in this set
[#0] ERROR:InputArguments -- RooArgSet::checkForDup: ERROR argument with name model_MOMENT_2C_mT is already in this set
[#1] INFO:NumericIntegration -- RooRealIntegral::init(model_MOMENT_2C_mPhi_product_Int[mPhi,mT]_Norm[mPhi,mT]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(mPhi,mT)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(model_MOMENT_2C_mT_product_Int[mPhi,mT]_Norm[mPhi,mT]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(mT,mPhi)
[#0] ERROR:InputArguments -- RooArgSet::checkForDup: ERROR argument with name model_MOMENT_2C_mPhi is already in this set
[#0] ERROR:InputArguments -- RooArgSet::checkForDup: ERROR argument with name model_MOMENT_2C_mT is already in this set
Info in <TCanvas::Print>: pdf file TEST.pdf has been created
and, interestingly, when I run Print("T") on the morphed PDF, I get the error
[#0] ERROR:InputArguments -- RooLinearVar::RooLinearVar(morph_func_transVar_0_0): ERROR, slope(morph_func_slope_0_0) and offset(morph_func_offset_0_0) may not depend on variable(mPhi)
which crashes the script. I’m not too sure how to interpret this, but since I’m trying to perform the interpolation along the variable mPhi, the fact that the morphed PDF may not depend on this variable is troubling!
I know this is slightly off-topic from my main post, and that the RooMomentMorphFuncND.Grid2 and RooMomentMorphFuncND classes in general are new and not very well-documented, but if you or perhaps Jonas (absolutely no rush) have any insights, I would greatly appreciate it.
I’ve attached the files needed to run the reproducible example, and the script can be run via
python InterpolateShapes.py -m 150 -m1 125 -m2 175 -mT 1800 -y 17
1800-125_17_ws.root (9.7 KB)
1800-175_17_ws.root (9.7 KB)
InterpolateShapes.py (5.1 KB)
Thanks a lot!