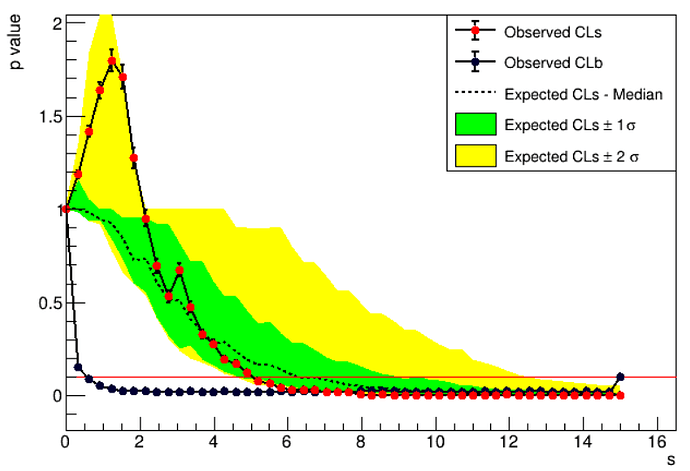
I create the Possion model with gaussian constrain backgrounds, I use the model to calculate the upper limit.
The model config is below:
#include <iostream>
#include "TFile.h"
#include "RooWorkspace.h"
#include "RooAbsPdf.h"
#include "RooRealVar.h"
#include "RooDataSet.h"
#include "RooStats/ModelConfig.h"
#include "RooRandom.h"
#include "TGraphErrors.h"
#include "TGraphAsymmErrors.h"
#include "TCanvas.h"
#include "TLine.h"
#include "TROOT.h"
#include "RooStats/AsymptoticCalculator.h"
#include "RooStats/HybridCalculator.h"
#include "RooStats/FrequentistCalculator.h"
#include "RooStats/ToyMCSampler.h"
#include "RooStats/HypoTestPlot.h"
#include "RooStats/NumEventsTestStat.h"
#include "RooStats/ProfileLikelihoodTestStat.h"
#include "RooStats/SimpleLikelihoodRatioTestStat.h"
#include "RooStats/RatioOfProfiledLikelihoodsTestStat.h"
#include "RooStats/MaxLikelihoodEstimateTestStat.h"
#include "RooStats/NumEventsTestStat.h"
#include "RooStats/HypoTestInverter.h"
#include "RooStats/HypoTestInverterResult.h"
#include "RooStats/HypoTestInverterPlot.h"
using namespace RooFit;
using namespace RooStats;
using namespace RooFit;
using namespace RooStats;
using namespace TMath;
using namespace std;
void PoissonModelWithGaussBkg(int nobs = 0, double eff0 = 1,double sigma_eff = 0.3, double b0 = 4.1, double rel_sigmab = 0.17) {
RooWorkspace w("w",true);
// make sum of s+b
w.factory("prod:effs(s[3,0,15],eff[1e-7,2.0])");
w.factory("sum:nexp(effs,b[0,15])");
//w.factory("sum:nexp(s[0],b[0,15])");
// Poisson of (n | s+b)
w.factory("Poisson:pdf(nobs[0,50],nexp)");
w.factory("Gaussian:constraint_eff(eff, eff0[1], sigma_eff[0.3])") ;
w.factory("Gaussian:constraint_bkg(b,b0[4.1],sigmab[4.1*0.17])");
w.factory("PROD:model(pdf,constraint_bkg,constraint_eff)");
RooRealVar * obs = w.var("nobs");
w.var("b0")->setVal(b0);
w.var("sigmab")->setVal(b0*rel_sigmab);
w.var("eff0")->setVal(eff0);
w.var("sigma_eff")->setVal(sigma_eff);
//set value of observed events
obs->setVal(nobs);
// Q: how to calculate the value ?
//w.var("s")->setVal(0.0);
//RooAbsPdf *model = w.pdf("model");
//RooAbsReal* integral = model->createIntegral(RooArgSet(*w.var("b"), *w.var("eff")));
//double prob = integral->getVal();
//cout <<"prob = "<<prob <<endl ;
// make data set with the namber of observed events
RooDataSet data("data","", *obs );
data.add(*obs );
w.import(data);
w.Print();
cout << "nobs = " << nobs << endl;
cout << "b0 = " << b0 << endl;
cout << "sigmab = " << b0*rel_sigmab << endl;
cout << "eff0 = " << eff0 << endl;
cout << "sigma_eff = " << sigma_eff << endl;
ModelConfig mc("ModelConfig",&w);
mc.SetPdf(*w.pdf("model"));
mc.SetParametersOfInterest(*w.var("s"));
mc.SetObservables(*w.var("nobs"));
// set an empty set since there are no nuisance parameters
mc.SetNuisanceParameters(RooArgSet(*w.var("b"),*w.var("eff")));
//mc.SetNuisanceParameters(RooArgSet(*w.var("b")));
// these are needed for the hypothesis tests
mc.SetSnapshot(*w.var("s"));
mc.SetGlobalObservables(*w.var("b0"));
mc.SetGlobalObservables(*w.var("sigmab"));
mc.SetGlobalObservables(*w.var("eff0"));
mc.SetGlobalObservables(*w.var("sigma_eff"));
mc.Print();
// import model in the workspace
w.import(mc);
TString fileName = TString::Format("data/PoissonModelWithGaussBkg_n%d.root",nobs);
// write workspace in the file (recreate file if already existing)
w.writeToFile(fileName, true);
cout << "model written to file " << fileName << endl;
}
and the hopytest Inv code is below: I use the hybridCalculator and use CLs to calculate the two side upperlimit.
#include "TFile.h"
#include "RooWorkspace.h"
#include "RooAbsPdf.h"
#include "RooRealVar.h"
#include "RooDataSet.h"
#include "RooStats/ModelConfig.h"
#include "RooRandom.h"
#include "TGraphErrors.h"
#include "TGraphAsymmErrors.h"
#include "TCanvas.h"
#include "TLine.h"
#include "TROOT.h"
#include "RooStats/AsymptoticCalculator.h"
#include "RooStats/HybridCalculator.h"
#include "RooStats/FrequentistCalculator.h"
#include "RooStats/ToyMCSampler.h"
#include "RooStats/HypoTestPlot.h"
#include "RooStats/NumEventsTestStat.h"
#include "RooStats/ProfileLikelihoodTestStat.h"
#include "RooStats/SimpleLikelihoodRatioTestStat.h"
#include "RooStats/RatioOfProfiledLikelihoodsTestStat.h"
#include "RooStats/MaxLikelihoodEstimateTestStat.h"
#include "RooStats/NumEventsTestStat.h"
#include "RooStats/HypoTestInverter.h"
#include "RooStats/HypoTestInverterResult.h"
#include "RooStats/HypoTestInverterPlot.h"
using namespace RooFit;
using namespace RooStats;
// internal routine to run the inverter
void HypoTestInvForLognormal(const char * infile = "",
const char* output_txt_name = "output.txt",
bool draw_flag = false,
int ToyNumEvent = 20000,
const char* workspaceName = "w",
const char* modelSBName = "ModelConfig",
const char* modelBName = "",
const char* dataName = "data" )
{
TString fileName(infile);
// Check if example input file exists
TFile *file = TFile::Open(fileName);
// if input file was specified but not found, quit
if(!file ) {
cout <<"file " << fileName << " not found" << endl;
return;
}
// get the workspace out of the file
RooWorkspace* w = (RooWorkspace*) file->Get(workspaceName);
if(!w){
cout <<"workspace not found" << endl;
return;
}
RooAbsData * data = w->data(dataName);
if (!data) {
Error("StandardHypoTestDemo","Not existing data %s",dataName);
w->Print();
return;
}
// check the model taken from the workspace
ModelConfig* bModel = (ModelConfig*) w->obj(modelBName);
ModelConfig* sbModel = (ModelConfig*) w->obj(modelSBName);
if (!sbModel) {
Error("StandardHypoTestDemo","Not existing ModelConfig %s",modelSBName);
w->Print();
return;
}
// check the model
if (!sbModel->GetPdf()) {
Error("StandardHypoTestDemo","Model %s has no pdf ",modelSBName);
w->Print();
return;
}
if (!sbModel->GetParametersOfInterest() || sbModel->GetParametersOfInterest()->getSize() == 0) {
Error("StandardHypoTestDemo","Model %s has no poi ",modelSBName);
w->Print();
return;
}
if (!sbModel->GetObservables() || sbModel->GetObservables()->getSize() == 0) {
Error("StandardHypoTestInvDemo","Model %s has no observables ",modelSBName);
w->Print();
return;
}
// assume exists only one parameter of interest
RooRealVar* poi = (RooRealVar*) sbModel->GetParametersOfInterest()->first();
if (!sbModel->GetSnapshot() ) {
Info("StandardHypoTestInvDemo","Model %s has no snapshot - make one using model poi",modelSBName);
sbModel->SetSnapshot( *poi );
}
if (!bModel || bModel == sbModel) {
Info("StandardHypoTestInvDemo","The background model %s does not exist",modelBName);
Info("StandardHypoTestInvDemo","Copy it from ModelConfig %s and set POI to zero",modelSBName);
bModel = (ModelConfig*) sbModel->Clone();
bModel->SetName(TString(modelSBName)+TString("_with_poi_0"));
double oldval = poi->getVal();
poi->setVal(0);
bModel->SetSnapshot( *poi );
poi->setVal(oldval);
}
/////////////////////////////////////////////////////////////
// REAL Tutorial starts here
////////////////////////////////////////////////////////////
// build test statistics and hypotest calculators for running the inverter
// create the calculator
// create the HypoTest calculator class
// frequentist calculator
FrequentistCalculator * fc = new FrequentistCalculator(*data, *bModel, *sbModel);
fc->SetToys(ToyNumEvent, ToyNumEvent); // 1000 for null (S+B) , 50 for alt (B)
// asymptotic calculator
// AsymptoticCalculator * ac = new AsymptoticCalculator(*data, *bModel, *sbModel);
// ac->SetOneSided(true); // rof one-side tests (limits)
// AsymptoticCalculator::SetPrintLevel(-1);
// // Hybrid calculator
HybridCalculator * hc = new HybridCalculator(*data, *bModel, *sbModel);
hc->SetToys(ToyNumEvent, ToyNumEvent); // 1000 for null (S+B) , 50 for alt (B)
// check for nuisance prior pdf in case of nuisance parameters
if (sbModel->GetNuisanceParameters() && sbModel->GetNuisanceParameters()->getSize() > 0) {
// for hybrid need nnuisance pdf
RooAbsPdf * nuisPdf = RooStats::MakeNuisancePdf(*sbModel,"nuisancePdf_sbmodel");
hc->ForcePriorNuisanceAlt(*nuisPdf);
hc->ForcePriorNuisanceNull(*nuisPdf);
}
// create hypotest inverter
// passing the desired calculator
HypoTestInverter calc(*hc);
// set confidence level
calc.SetConfidenceLevel(0.90);
// for CLS
bool useCLs = true;
calc.UseCLs(useCLs);
calc.SetVerbose(true);
// configure ToyMCSampler
ToyMCSampler *toymcs = (ToyMCSampler*)calc.GetHypoTestCalculator()->GetTestStatSampler();
// for number counting (otherwise need to use extended pdf
toymcs->SetNEventsPerToy(1);
// profile likelihood test statistics
ProfileLikelihoodTestStat profll(*sbModel->GetPdf());
profll.SetOneSided(false);
// for CLs (bounded intervals) use one-sided profile likelihood
//if (useCLs) profll.SetOneSided(true);
// ratio of profile likelihood - need to pass snapshot for the alt
//RatioOfProfiledLikelihoodsTestStat ropl(*sbModel->GetPdf(), *bModel->GetPdf(), bModel->GetSnapshot());
// max likelihood
//MaxLikelihoodEstimateTestStat maxll(*sbModel->GetPdf(),*poi);
// number of events test statistics
NumEventsTestStat nevtts;
// set the test statistic to use
toymcs->SetTestStatistic(&profll);
//toymcs->SetTestStatistic(&ropl);
//toymcs->SetTestStatistic(&nevtts);
//toymcs->SetTestStatistic(&maxll);
// control message
//RooMsgService::instance().getStream(1).removeTopic(RooFit::NumIntegration);
// suppress info and progress messages
//RooMsgService::instance().setGlobalKillBelow(RooFit::WARNING) ;
int npoints = 50; // number of points to scan
// min and max (better to choose smaller intervals)
double poimin = poi->getMin();
double poimax = poi->getMax();
//poimin = -2; poimax=4;
std::cout << "Doing a fixed scan in interval : " << poimin << " , " << poimax << std::endl;
calc.SetFixedScan(npoints,poimin,poimax);
HypoTestInverterResult * r = calc.GetInterval();
// analyze result produced by the inverter
double upperLimit = r->UpperLimit();
double ulError = r->UpperLimitEstimatedError();
// double lowerLimit = r->LowerLimit();
// double llError = r->LowerLimitEstimatedError();
// if (lowerLimit < upperLimit*(1.- 1.E-4))
// std::cout << "The computed lower limit is: " << lowerLimit << " +/- " << llError << std::endl;
std::cout << "The computed upper limit is: " << upperLimit << " +/- " << ulError << std::endl;
// compute expected limit
std::cout << "Expected upper limits, using the B (alternate) model : " << std::endl;
std::cout << " expected limit (median) " << r->GetExpectedUpperLimit(0) << std::endl;
std::cout << " expected limit (-1 sig) " << r->GetExpectedUpperLimit(-1) << std::endl;
std::cout << " expected limit (+1 sig) " << r->GetExpectedUpperLimit(1) << std::endl;
//output the result
const RooDataSet* dataSet = dynamic_cast<RooDataSet*>(data);
int nobs = -1;
if (dataSet) {
for (int i = 0; i < dataSet->numEntries(); ++i) {
const RooArgSet* dataPoint = dataSet->get(i);
RooRealVar* x = dynamic_cast<RooRealVar*>(dataPoint->find("nobs"));
if (x) {
double xValue = x->getVal();
nobs = int(xValue) ;
}
}
}
std::ofstream outputFile(output_txt_name, std::ios_base::app);
if (outputFile.is_open()) {
outputFile << nobs << " "<< upperLimit << std::endl;
outputFile.close();
std::cout << "Upper Limit save to output.txt successfully" << std::endl;
} else {
std::cerr << "can't open output.txt。" << std::endl;
}
// plot now the result of the scan
if(draw_flag){
std::string strFilename(infile);
size_t pos = strFilename.find_last_of(".");
std::string substring = strFilename.substr(0, pos);
std::stringstream ss;
ss << substring << "_CLs_scan";
std::string filename = ss.str();
HypoTestInverterPlot *plot = new HypoTestInverterPlot("HTI_Result_Plot","Feldman-Cousins Interval",r);
// plot in a new canvas with style
TCanvas * c1 = new TCanvas("HypoTestInverter Scan");
c1->SetLogy(false);
plot->Draw("CLb"); // plot also CLb and CLs+b
//plot->Draw("OBS"); // plot only observed p-value
// plot also in a new canvas the test statistics distributions
// plot test statistics distributions for the two hypothesis
// when distribution is generated (case of FrequentistCalculators)
c1->SaveAs((filename + ".png").c_str());
//const int n = r->ArraySize();
//if (n> 0 && r->GetResult(0)->GetNullDistribution() ) {
// TCanvas * c2 = new TCanvas("Test Statistic Distributions","",2);
// if (n > 1) {
// int ny = TMath::CeilNint( sqrt(n) );
// int nx = TMath::CeilNint(double(n)/ny);
// c2->Divide( nx,ny);
// }
// for (int i=0; i<n; i++) {
// if (n > 1) c2->cd(i+1);
// SamplingDistPlot * pl = plot->MakeTestStatPlot(i);
// pl->SetLogYaxis(true);
// pl->Draw();
// }
//}
}
// optionally save result in a file
// TFile * fileOut = new TFile(mResultFileName,"RECREATE");
// r->Write();
// fileOut->Close();
delete r;
delete hc;
}
I have question:
1 , How do I check the model is right or not? The code is in high level and I don’t know how to check the model I created
- For the Poission model, I also want to set s= 0, and convolute the backgrounds b and then give the nobs distribution. So anyone can help me how to do this things? Thank you .