Hi all,
I am having some problems doing unbinned ML Fit in regions. I have data in range [70,120] of which [86,96] is my blind region (“blind”) and both side of it are the sidebands ("unblindReg_1 , unblindReg_2). The integral of data in [70,120] is 2637 ( blind: 614 and sidebands:2023). I am using a power law as my model(RooGenericPdf). I need to fit my model in the side bands → extract the parameter value (p) → Find normalisation in blind region from the model with parameter value p.
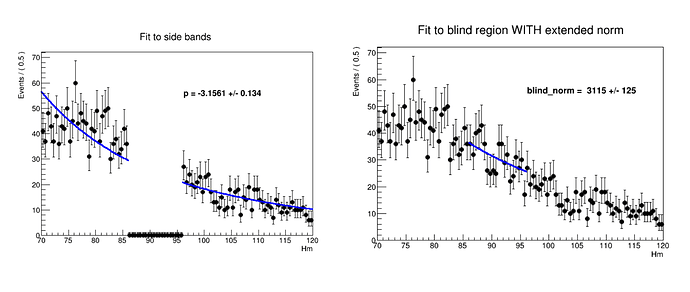
1.First I fit the model to data sideband regions without any extended normalisation going into the fit. The fit looks fine.
2. Take a model with parameter value same as obtained from step1 and extend it with a norm parameter → fitTo data in blind region → obtain the normalisation. Here the normalisation is coming out to be 3115 , even greater than integral of data in entire range!
I have attached the plots and the code snippet. It would be really helpful to know what i am missing here or is there a better way to do what i am trying to do.
###Step1: Fiting in sidebands without an extended norm.
p = r.RooRealVar(“p”,“p”,-1,-5,0)
data_bkg = r.RooGenericPdf(“data_bkg”,“data_bkg”,“Hm^p”,r.RooArgList(Hm,p))
fitresult = data_bkg.fitTo(data_obs,r.RooFit.Range(“unblindReg_1,unblindReg_2”),r.RooFit.Save())
###Step2:Fiting in blind region with parameter from side band fits with an extended norm.
p = r.RooRealVar(“p”,“p”,-3.1562)
data_bkg2 = r.RooGenericPdf(“data_bkg2”,“data_bkg2”,“Hm^p”,r.RooArgList(Hm,p))
blind_norm = r.RooRealVar(“blind_norm”,“blind_norm”,500,0,3500)
data_bkg2_=r.RooAddPdf(“data_bkg2_”,“data_bkg2_”,r.RooArgList(data_bkg2),r.RooArgList(blind_norm))
fitresult = data_bkg2_.fitTo(data_obs,r.RooFit.Range(“blind”),r.RooFit.Save())
Thanks,
Sweta