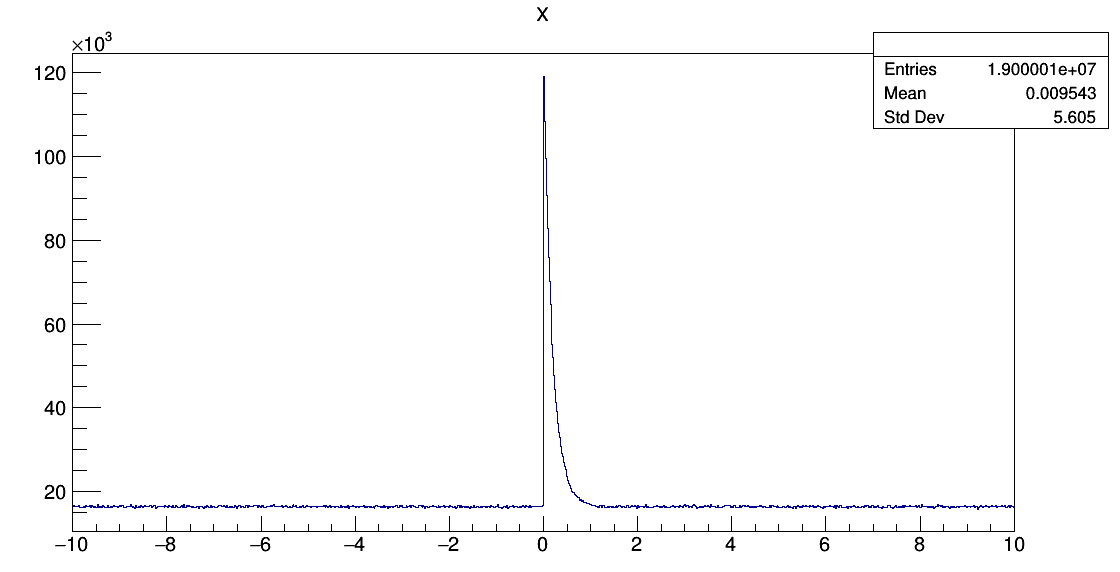
Dear Ganis,
Thank you very much for your prompt reply.
Following your suggestion, I fixed the constant background by fitting the negative range [-10,0] with a extended Polynomial model to derive mean value and error of the background. Then I used those values to constrain the background in the (extended) combined model of background and exponential functions. Here is the code:
#include "RooRealVar.h"
#include "RooDataSet.h"
#include "RooGaussian.h"
#include "RooArgSet.h"
#include "RooExponential.h"
#include "RooChebychev.h"
#include "RooPolynomial.h"
#include "RooAddPdf.h"
#include "RooExtendPdf.h"
#include "TCanvas.h"
#include "TAxis.h"
#include "RooPlot.h"
#include "TFile.h"
#include "TTree.h"
#include "fitF.cxx"
using namespace RooFit ;
void fitdev()
{
//! parameters
Double_t p_timerange=10.;
Double_t p_bkgmaxerr=1000.;
Double_t p_sigmaxerr=10000.;
//! Input data
TFile *f=TFile::Open("sim_mlh.root");
TTree* tree;
f->GetObject("treeb",tree);
//! initial guess
// Calculate number of backgrounds
Double_t guessbkg=(Double_t)tree->Draw("",Form("x>%f&&x<0",-p_timerange),"goff");
// Calculate number of signals
Double_t guesssig=(Double_t)tree->Draw("",Form("x<%f&&x>0",p_timerange),"goff")-guessbkg;
//! Fitting Negative background
RooRealVar xn("x","x",-10.,0.) ;
RooPolynomial bkgn("bkgn","bkgd negative pdf",xn);
RooRealVar nbkgn("nbkgn","number of negative background events",guessbkg,guessbkg-p_bkgmaxerr,guessbkg+p_bkgmaxerr) ;
RooExtendPdf ebkgn("ebkgn","extended negative background p.d.f",bkgn,nbkgn) ;
RooDataSet* datan=new RooDataSet("data","data set",tree,xn);
ebkgn.fitTo(*datan);
ebkgn.Print("t") ;
cout<<"\n\n*******Finished fitting background******* \n"<<endl;
//! Fitting positive range
RooRealVar x("x","x",0.,10.) ;
RooRealVar hl("hl","half life",0.1,0.01,10.);
RooPolynomial bkg("bkg","bkgd pdf",x);
fitF sig("sig","sig pdf",x,hl);
RooRealVar nsig("nsig","number of signal events",guesssig,guesssig-p_sigmaxerr,guesssig+p_sigmaxerr) ;
RooRealVar nbkg("nbkg","number of background events",guessbkg,guessbkg-p_bkgmaxerr,guessbkg+p_bkgmaxerr) ;
RooExtendPdf ebkg("ebkg","extended background p.d.f",bkg,nbkg) ;
RooExtendPdf esig("esig","extended signal p.d.f",sig,nsig) ;
RooAddPdf model("model","(g1+g2)+a",RooArgList(ebkg,esig)) ;
RooDataSet* data=new RooDataSet("data","data set",tree,x);
// Fit with constrain on background derived from negative part
RooGaussian constrlBkg("constrainBkg","constrainBkg",nbkg, RooConst(nbkgn.getVal()), RooConst(nbkgn.getError()));
RooArgSet constrset;
constrset.add(constrlBkg);
model.fitTo(*data,ExternalConstraints(constrset));
model.Print("t") ;
//! Plotting
RooPlot* xframe = x.frame(Title("extended ML fit example")) ;
data->plotOn(xframe) ;
model.plotOn(xframe,Normalization(1.0,RooAbsReal::RelativeExpected)) ;
new TCanvas("rf202_composite","rf202_composite",600,600) ;
gPad->SetLeftMargin(0.15) ; xframe->GetYaxis()->SetTitleOffset(1.4) ; xframe->Draw() ;
}
Where I defined the exponential function (for further developement) as follow:
/*****************************************************************************
* Project: RooFit *
* *
* This code was autogenerated by RooClassFactory *
*****************************************************************************/
// Your description goes here...
#include "Riostream.h"
#include "fitF.h"
#include "RooAbsReal.h"
#include "RooAbsCategory.h"
#include <math.h>
#include "TMath.h"
ClassImp(fitF)
fitF::fitF(const char *name, const char *title,
RooAbsReal& _x,
RooAbsReal& _hl
) :
RooAbsPdf(name,title),
x("x","x",this,_x),
hl("hl","hl",this,_hl)
{
}
fitF::fitF(const fitF& other, const char* name) :
RooAbsPdf(other,name),
x("x",this,other.x),
hl("hl",this,other.hl)
{
}
Double_t fitF::evaluate() const
{
// ENTER EXPRESSION IN TERMS OF VARIABLE ARGUMENTS HERE
return exp(-log(2)/hl*x);
}
The result is in good agreement with the value from simulation.
Do you think this method is correct?
Thank again!
Phong