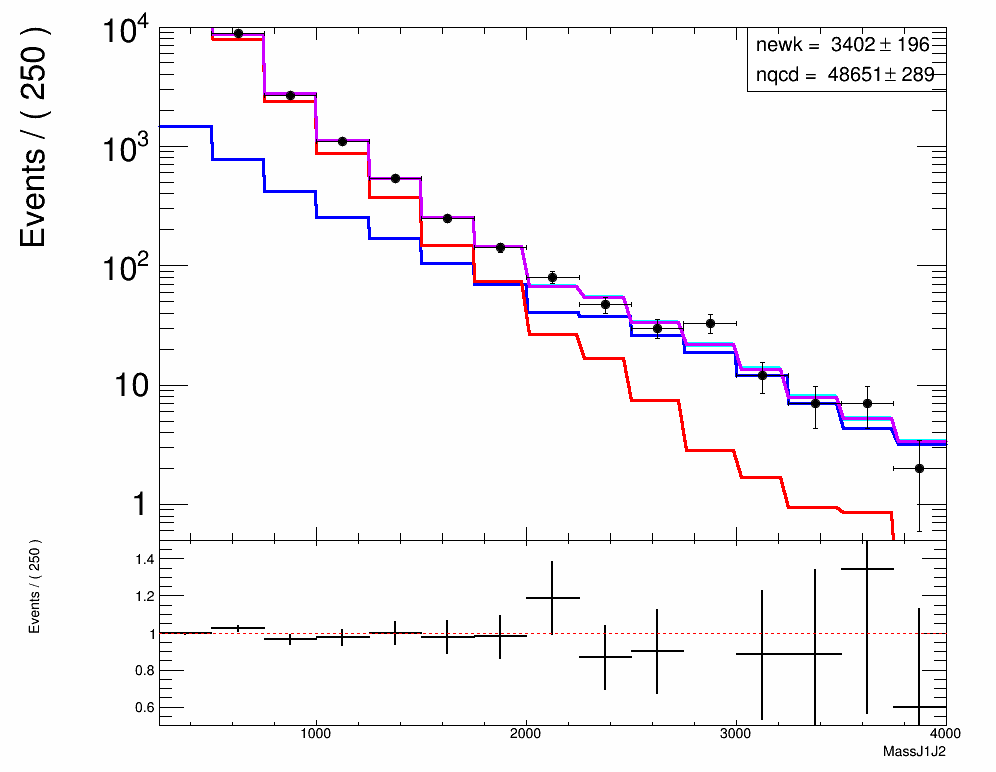
Hi All,
I am trying to do an extended binned maximum likelihood fit in order to extract the relative number of signal and background events in data. I seem to have a working set-up but when I try to plot the RooAddPdf components, there are displayed with weird shaped bins.
Please find the relevant code snippet below. hData, hQCD and hEWK are TH1Fs.
ndata = hData.Integral()
fitRangeMin = 250.0
fitRangeMax = 4000.0
# Define fit variable axis
varX = RooRealVar("varX",hData.GetXaxis().GetTitle(),fitRangeMin,fitRangeMax)
# Define variables for fitted signal and QCD events
newk = RooRealVar("newk","number of events in the EWK signal",ndata*1./10., 1, ndata)
nqcd = RooRealVar("nqcd","number of events in the QCD background",ndata*9./10., 1, ndata)
# Get the histograms for RooFit
data = RooDataHist("data" ,"data", RooArgList(varX), hData)
ewk = RooDataHist( "ewk" ,"EWK" , RooArgList(varX), hEWK)
qcd = RooDataHist( "qcd" ,"QCD" , RooArgList(varX), hQCD)
# Set the histograms for ewk and qcd as PDFs
ewkModel = RooHistPdf("ewkModel","RooFit template for EWK signal", RooArgSet(varX), ewk)
qcdModel = RooHistPdf("qcdModel","RooFit template for QCD backgrounds" , RooArgSet(varX), qcd)
# Prepare extended likelihood fit
ewkShape = RooExtendPdf("ewkShape","EWK shape pdf", ewkModel, newk)
qcdShape = RooExtendPdf("qcdShape","QCD shape pdf", qcdModel, nqcd)
# Combine the models
combModel = RooAddPdf("combModel","Combined model for EWK and QCD", RooArgList(ewkShape,qcdShape))
# Fit - options provided are
# RooFit.Extended(True) -> do extended likelihood fit
# RooFit.Save(True) -> save RooFitResult object
# RooFit.Minos(True) -> Call MINOS after HESSE, itself after MIGRAD
fitResult = combModel.fitTo(data, RooFit.Extended(True), RooFit.Save(True), RooFit.Minos(True))
can2 = makeCanvas('c2')
can2.SetLogy()
can2.cd()
can2.SetLeftMargin(0.15)
up2 = ROOT.TPad("mypad1", "mypad1", 0.0, 0.30, 1.0, 1.00)
up2.SetBottomMargin(0)
up2.SetLogy()
up2.Draw()
down2 = ROOT.TPad("mypad2", "mypad2", 0.0, 0.00, 1.0, 0.30)
down2.SetTopMargin(0)
down2.SetBottomMargin(0.2)
down2.Draw()
up2.cd()
# Set up the plotting with RooFit
xframe = varX.frame()
#option tells plotOn to not take number density but number in histogram
data.plotOn(xframe, RooFit.DataError(RooAbsData.SumW2))
combModel.plotOn(xframe, RooFit.VisualizeError(fitResult))
combModel.plotOn(xframe, RooFit.Components("ewkShape"), RooFit.LineColor(ROOT.kBlue))
combModel.plotOn(xframe, RooFit.Components("qcdShape"), RooFit.LineColor(ROOT.kRed))
combModel.plotOn(xframe, RooFit.LineColor(ROOT.kViolet))
data.plotOn(xframe, RooFit.DataError(RooAbsData.SumW2)) #Data lies over all else, check that same as input TH1
xframe.SetMaximum(1e4) #calls TH1::SetMaximum, helps with log scale
xframe.SetMinimum(0.5)
combModel.paramOn(xframe,RooFit.Layout(0.75,0.95,0.95))
xframe.Draw()
down2.cd()
ROOT.gStyle.SetOptStat(0)
xframedown = varX.frame()
# Get fitted histogram by calculating number of bins manually
nbins = (fitRangeMax-fitRangeMin)/hData.GetXaxis().GetBinWidth(1)
hFitted = combModel.createHistogram("fitted_",varX,RooFit.Binning(int(nbins),fitRangeMin,fitRangeMax))
#Scale with fit results, since PDFs are normalized to something-FIGURE OUT
# print 'CHECK -', hFitted.Integral()
integralBefore = hFitted.Integral()
hFitted.Scale((nqcd.getVal()+newk.getVal())/integralBefore)
# print 'CHECK -', hFitted.Integral()
hDataAfter = data.createHistogram("data_",varX,RooFit.Binning(int(nbins),fitRangeMin,fitRangeMax))
# hDataAfter.SetLineColor(ROOT.kViolet)
# hDataAfter.Draw("EP SAME")
# hFitted.SetLineColor(ROOT.kGreen)
# hFitted.SetLineStyle(2)
# hFitted.Draw("HIST SAME")
hResidual = hDataAfter.Clone('DataAfter')
hResidual.Divide(hFitted)
hResidual.SetMaximum(1.5)
hResidual.SetMinimum(0.5)
hResidual.SetStats(False)
hResidual.Draw()
# xmin2 = up2.GetUxmin()
# xmax2 = up2.GetUxmax()
xmin2 = fitRangeMin
xmax2 = fitRangeMax
line2 = ROOT.TLine(xmin2,1,xmax2,1)
line2.SetLineColor(ROOT.kRed)
line2.SetLineStyle(2)
line2.Draw('SAME')
print 'Fitted # EWK: ',newk.getVal(), newk.getError()
print 'Fitted # EWK: ',newk.Print()
fitResult.Print()
Could you please tell me what I am doing wrong?
Thanks!
-Yaadav