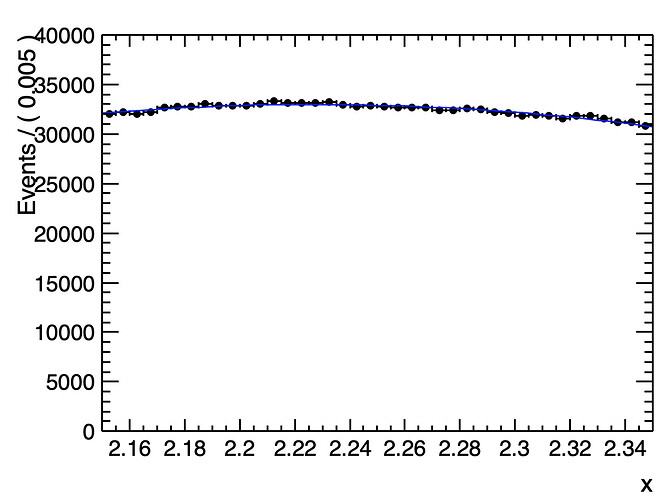
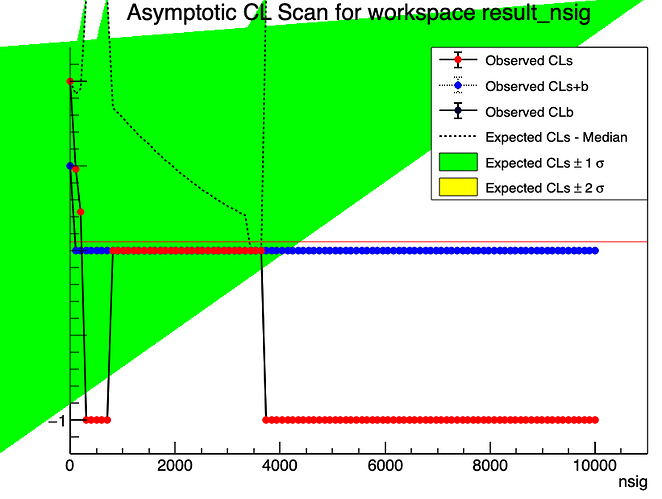
I am doing a counting experiment, looking for a particle using invariant mass analysis. I have considerable background but see no visible peak/signal. What I need to do is set an upper limit to the signal, but I am unsure which method is the best to use for my analysis. I have used ProfileLikelihoodCalculator, which gives me a number for upper limit, this is fine.
I am trying to use StandardHypoTestInvDemo.c to also compute a limit, but it’s a bit more complicated. There are a lot more inputs that I am unaware of which setting to use (calculatorType, teststatType, npoints, ntoys…). My question seems to be, which should be the better way to calculate this upper limit and which settings should I be using? I can mess around with them but i’ll really never know if I did it correctly.
If uploaded correctly, the screenshot should have the black data points with a blue fit through them. The blue fit is a 2nd degree polynomial with a briet-wigner from 2.245 to 2.265, mean of 2.255 and width of .01. Since there is no visible signal, the background clearly dominates.
Any resources that would help me are greatly appreciated. I can also share my code and .root file if necessary. Thank you!