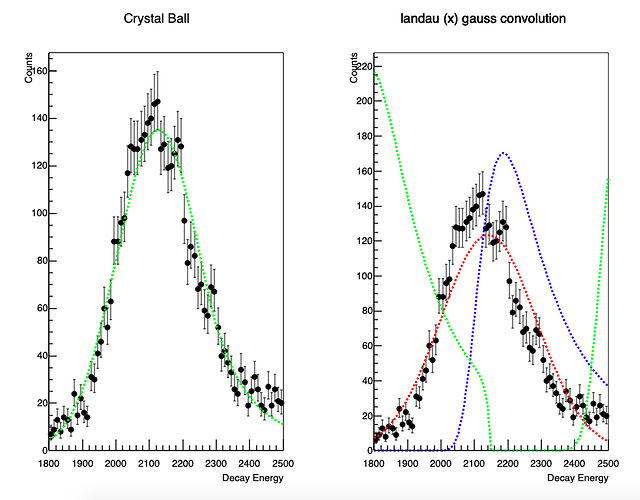
Hello,
I have a histogram, which has different proton peaks- which are basically broad distributions. The high energy tailing in the peaks is due to the energy deposited by the betas in addition to the protons which are detected in the DSSD detector.
dssd-fit.pdf (37.7 KB)
Each peak visible in the histogram is actually a sum of two or three peaks, with known relative intensities.
For eg. The first dominant peak is sum of three peaks with supposed centroids at
1685 keV ->1878 channel (4% rel intensity)
1836 keV ->2029 channel ( 36% rel intensity)
1902 keV -> 2095 channel (100% rel intensity)
Im fitting this structure, as a total of three gaussians. But, I am unable to constrain the fits and reproduce the correct intensities. Also I end up having bad chi_square value.
The way I define the fitting routine is as follows -
xpos[0]=1878.0;
xpos[1]=2029.0;
xpos[2]=2095.0;
ypos[0] = decayEnergy->GetBinContent(xpos[0]);
ypos[1] = decayEnergy->GetBinContent(xpos[1]);
ypos[2] = decayEnergy->GetBinContent(xpos[2]);
TCanvas *cEnergy = new TCanvas("cEnergy","cEnergy", 800,600);
cEnergy->cd();
cout<<"Ypos "<<ypos[0]<<endl;
TF1 * F1 = new TF1("F1", "gaus",1446,2310); // Ep = 1685 peak -> 1878 channel# (3sigma= 144*3 = 432)
TF1 * F2 = new TF1("F2", "gaus",1597,2461); // Ep = 1838 peak ->2029 channel
TF1 * F3 = new TF1("F3", "gaus",1663,2527);// Ep = 1902 peak -> 2095 channel
TF1 * total = new TF1("total", "gaus(0)+gaus(3)+gaus(6)",1440,2527);
F1->FixParameter(0,ypos[0]);
F1->FixParameter(1,1878);
F1->SetParameter(2,70);
F1->SetLineColor(kGreen);
// F1->FixParameter(0,90.0);
decayEnergy->Fit("F1","SR");
F2->FixParameter(0,ypos[1]);
F2->FixParameter(1,2029);
F2->SetParameter(2,70);
F2->SetLineColor(kBlack);
// F2->FixParameter(0,300.0);
decayEnergy->Fit("F2","SR+");
F3->FixParameter(0,ypos[2]);
F3->SetParameter(1,2095);
F3->FixParameter(2,70);
F3->SetLineColor(kRed);
// F3->FixParameter(0,450.0);
decayEnergy->Fit("F3","SER+");
F1->GetParameters(&par[0]);
F2->GetParameters(&par[3]);
F3->GetParameters(&par[6]);
total->SetParameters(par);
total->SetParLimits(1,1878,1920); // centroids
total->SetParLimits(4,2030,2070);
total->SetParLimits(7,2090,2130);
total->SetParLimits(0,1,50); // heights
total->SetParLimits(3,1,200);
total->SetParLimits(6,1,200);
total->SetParLimits(2,70,150); // sigmas
total->SetParLimits(5,70,150);
total->SetParLimits(8,70,150);
total->SetLineColor(kMagenta);
decayEnergy ->Fit("total", "SR+");
total->Draw(" same");
Is there any better way to do this routine. Or is some other routine available within ROOT framework, which can be used
Thanks
Mansi