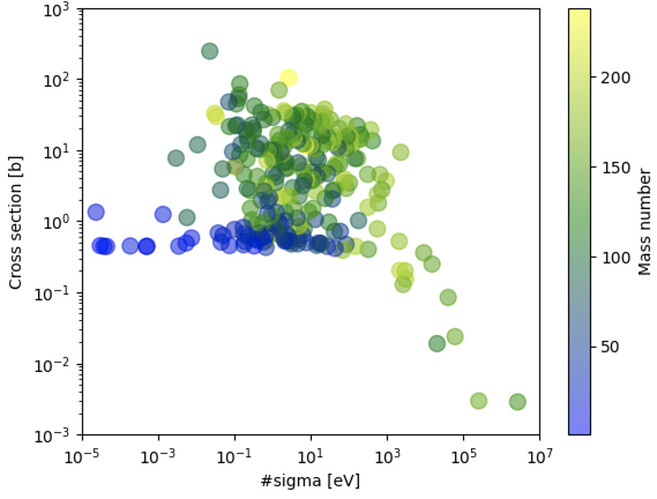
I have a TGraph that its point represents two nuclear properties for different nucleus.
I would like to create a TH2Poly and weigh the points by a 3rd nuclear property.
I create the TGraph from the first two columns (1st olumn is x and 2nd is y) from an ascii file that looks like that
2.650e+006 2.868e-003 135.00
2.540e+005 2.969e-003 157.00
6.090e+004 2.376e-002 155.00
4.014e+004 8.445e-002 149.00
2.060e+004 1.893e-002 113.00
1.517e+004 2.482e-001 151.00
9.200e+003 3.587e-001 151.00
3.080e+003 1.532e-001 196.00
3.000e+003 2.003e-001 184.00
2.650e+003 1.287e-001 164.00
2.300e+003 9.261e+000 168.00
2.150e+003 2.023e-001 199.00
2.090e+003 5.201e-001 176.00
9.540e+002 3.669e+000 191.00
7.350e+002 2.748e+000 152.00
6.590e+002 4.507e+000 167.00
6.000e+002 1.792e+000 161.00
5.630e+002 2.396e+000 180.00
5.610e+002 7.772e-001 174.00
4.180e+002 1.347e+001 123.00
3.730e+002 1.923e+001 177.00
3.250e+002 4.000e-001 143.00
3.200e+002 1.563e+000 187.00
3.120e+002 4.551e+000 153.00
2.060e+002 1.442e+001 152.00
2.020e+002 1.634e+001 115.00
1.940e+002 1.412e+001 162.00
1.800e+002 1.017e+000 83.00
1.684e+002 7.565e+000 147.00
1.650e+002 2.182e+001 124.00
1.520e+002 4.408e-001 190.00
1.450e+002 7.586e+000 103.00
1.240e+002 1.185e+001 163.00
1.120e+002 1.533e+001 185.00
1.110e+002 1.216e+001 193.00
1.050e+002 1.638e+001 169.00
1.040e+002 3.442e+000 150.00
9.865e+001 1.571e+001 197.00
9.100e+001 1.538e+001 109.00
8.500e+001 2.882e+000 154.00
8.500e+001 4.706e-001 76.00
8.500e+001 1.059e+001 131.00
8.400e+001 2.321e+001 178.00
8.000e+001 3.500e+000 186.00
7.640e+001 3.927e+000 187.00
6.940e+001 3.890e-001 174.00
6.470e+001 1.028e+001 165.00
6.000e+001 7.167e-001 50.00
5.720e+001 6.329e+000 138.00
5.700e+001 1.360e+001 147.00
5.600e+001 2.004e+001 160.00
5.180e+001 1.081e+001 74.00
4.860e+001 6.481e+000 171.00
4.360e+001 4.128e-001 35.00
4.300e+001 2.791e+000 158.00
4.200e+001 5.714e+000 145.00
4.200e+001 7.143e-001 77.00
4.100e+001 1.537e+001 179.00
3.790e+001 1.280e+001 186.00
3.760e+001 2.660e+000 107.00
3.718e+001 2.041e+000 59.00
3.300e+001 2.679e+001 156.00
3.000e+001 9.667e-001 115.00
2.900e+001 1.507e+001 133.00
2.800e+001 4.643e+000 82.00
2.750e+001 1.327e+001 195.00
2.720e+001 4.412e-001 45.00
2.500e+001 2.696e+001 189.00
2.400e+001 2.083e+000 111.00
2.350e+001 3.745e+001 176.00
2.340e+001 1.786e+001 159.00
2.310e+001 2.641e+001 175.00
2.100e+001 1.190e+001 129.00
2.070e+001 2.918e+001 182.00
2.050e+001 3.220e+001 181.00
2.000e+001 1.600e+001 99.00
2.000e+001 4.900e+000 105.00
1.960e+001 4.847e+000 166.00
1.900e+001 2.526e+001 162.00
1.870e+001 4.813e-001 142.00
1.820e+001 4.890e-001 53.00
1.710e+001 2.222e+001 173.00
1.600e+001 7.313e+000 87.00
1.590e+001 4.906e-001 50.00
1.500e+001 4.267e+000 73.00
1.450e+001 4.552e-001 62.00
1.400e+001 7.929e+000 95.00
1.330e+001 1.053e+000 55.00
1.304e+001 2.684e+000 180.00
1.300e+001 1.031e+001 164.00
1.200e+001 2.583e+001 113.00
1.150e+001 4.878e+000 80.00
1.150e+001 1.513e+000 141.00
1.140e+001 2.570e+001 170.00
1.140e+001 3.772e+000 203.00
1.130e+001 1.504e+001 130.00
1.100e+001 3.727e+000 110.00
1.100e+001 1.155e+001 79.00
1.010e+001 3.337e+001 183.00
1.000e+001 1.150e+001 192.00
8.930e+000 1.355e+000 139.00
8.400e+000 4.286e+000 154.00
8.300e+000 2.940e+001 108.00
7.840e+000 4.974e-001 48.00
7.800e+000 3.846e+000 201.00
7.370e+000 1.153e+001 232.00
7.100e+000 2.254e+001 99.00
6.800e+000 7.794e-001 124.00
6.800e+000 3.676e+000 67.00
6.500e+000 5.077e+000 132.00
6.300e+000 1.222e+001 136.00
6.200e+000 6.339e-001 43.00
6.200e+000 3.145e+000 78.00
6.200e+000 2.419e+001 127.00
5.900e+000 3.475e+001 121.00
5.800e+000 6.103e+000 170.00
5.800e+000 1.672e+001 135.00
5.100e+000 8.431e-001 137.00
5.000e+000 2.240e+000 100.00
4.900e+000 5.510e-001 51.00
4.890e+000 8.589e-001 202.00
4.710e+000 6.624e+000 71.00
4.700e+000 3.234e+001 188.00
4.600e+000 4.783e-001 58.00
4.500e+000 1.356e+001 75.00
4.500e+000 1.104e+000 63.00
4.100e+000 3.098e+001 123.00
3.660e+000 1.475e+001 198.00
3.600e+000 1.500e+000 144.00
3.500e+000 1.714e+001 126.00
3.400e+000 2.618e+001 122.00
3.400e+000 2.941e+000 102.00
3.400e+000 2.941e+001 101.00
3.150e+000 4.762e-001 70.00
2.900e+000 5.172e-001 60.00
2.850e+000 2.211e+000 176.00
2.740e+000 1.350e+001 168.00
2.700e+000 1.852e+001 81.00
2.680e+000 1.034e+002 238.00
2.590e+000 5.405e-001 56.00
2.500e+000 6.000e-001 61.00
2.500e+000 5.600e+000 148.00
2.480e+000 6.452e-001 57.00
2.400e+000 1.458e+001 148.00
2.300e+000 6.826e+000 117.00
2.250e+000 5.333e-001 54.00
2.200e+000 3.318e+001 158.00
2.200e+000 1.318e+000 119.00
2.200e+000 5.455e-001 49.00
2.200e+000 5.455e+000 112.00
2.170e+000 1.009e+000 65.00
2.100e+000 6.667e+000 97.00
2.100e+000 5.238e-001 39.00
2.000e+000 3.550e+001 198.00
2.000e+000 2.300e+000 192.00
2.000e+000 1.100e+001 134.00
1.700e+000 8.647e+000 184.00
1.700e+000 8.824e-001 47.00
1.680e+000 9.286e+000 69.00
1.550e+000 1.355e+001 125.00
1.520e+000 6.447e-001 64.00
1.500e+000 6.933e+001 156.00
1.460e+000 9.589e-001 41.00
1.440e+000 2.153e+000 194.00
1.400e+000 1.657e+001 146.00
1.280e+000 7.813e-001 89.00
1.280e+000 1.328e+000 58.00
1.240e+000 4.194e+000 91.00
1.210e+000 3.471e+000 102.00
1.200e+000 1.167e+001 150.00
1.150e+000 7.391e+000 93.00
1.100e+000 1.000e+001 108.00
1.040e+000 7.212e+000 126.00
1.040e+000 4.615e+000 86.00
1.010e+000 2.871e+001 112.00
9.800e-001 8.163e-001 72.00
9.500e-001 1.211e+000 142.00
8.800e-001 6.591e-001 44.00
8.700e-001 9.885e+000 84.00
8.500e-001 2.118e+000 66.00
8.000e-001 3.125e+001 172.00
7.700e-001 9.610e+000 160.00
7.600e-001 6.579e-001 52.00
7.600e-001 1.908e+000 64.00
7.400e-001 1.270e+000 46.00
7.200e-001 7.083e+000 196.00
7.120e-001 5.478e-001 207.00
7.000e-001 3.400e+000 144.00
6.800e-001 4.265e-001 42.00
6.610e-001 3.026e+000 204.00
6.600e-001 6.212e-001 40.00
6.100e-001 2.623e+000 80.00
6.000e-001 2.667e+001 104.00
5.900e-001 5.085e-001 46.00
5.700e-001 9.474e-001 140.00
5.300e-001 5.868e-001 23.00
5.100e-001 1.961e+000 74.00
5.000e-001 3.400e+001 96.00
4.800e-001 1.229e+001 85.00
4.500e-001 1.022e+001 132.00
4.330e-001 6.813e-001 37.00
4.100e-001 5.366e-001 40.00
4.000e-001 4.250e+000 136.00
3.800e-001 1.053e+001 78.00
3.600e-001 8.889e-001 138.00
3.600e-001 5.556e-001 54.00
3.400e-001 4.147e+001 114.00
3.326e-001 4.480e-001 1.00
3.200e-001 2.000e+001 104.00
3.150e-001 1.810e+001 106.00
2.900e-001 1.034e+000 130.00
2.900e-001 2.241e+001 96.00
2.650e-001 1.509e+000 134.00
2.600e-001 2.692e+000 136.00
2.310e-001 6.061e-001 27.00
2.270e-001 1.366e+001 110.00
2.200e-001 1.545e+001 118.00
2.200e-001 2.864e+000 92.00
2.150e-001 7.442e+000 128.00
1.990e-001 1.910e+001 100.00
1.900e-001 5.158e-001 25.00
1.800e-001 4.500e+000 122.00
1.790e-001 6.592e-001 50.00
1.770e-001 4.633e-001 28.00
1.720e-001 8.140e-001 31.00
1.500e-001 1.200e+001 76.00
1.400e-001 8.571e+000 120.00
1.400e-001 8.500e+001 116.00
1.340e-001 5.970e+001 124.00
1.300e-001 5.308e+001 98.00
1.200e-001 2.250e+001 87.00
1.150e-001 4.435e+001 114.00
1.100e-001 2.182e+001 84.00
1.070e-001 5.888e+000 30.00
1.040e-001 5.865e+000 205.00
1.010e-001 7.624e-001 29.00
9.100e-002 9.451e+000 70.00
7.500e-002 2.133e+001 116.00
7.500e-002 4.533e-001 14.00
7.200e-002 4.722e+001 68.00
5.100e-002 6.275e-001 24.00
4.990e-002 5.411e+000 94.00
4.550e-002 5.055e-001 22.00
4.400e-002 2.727e+000 82.00
3.820e-002 6.806e-001 26.00
3.380e-002 2.959e+001 209.00
3.060e-002 3.269e+001 206.00
2.290e-002 2.445e+002 96.00
1.100e-002 1.182e+001 90.00
7.600e-003 5.789e-001 9.00
5.500e-003 4.909e-001 11.00
3.530e-003 4.448e-001 12.00
1.370e-003 1.241e+000 13.00
5.190e-004 4.432e-001 2.00
5.000e-004 4.400e-001 10.00
1.900e-004 4.474e-001 16.00
4.540e-005 4.405e-001 7.00
3.850e-005 4.416e-001 6.00
3.100e-005 4.516e-001 3.00
2.400e-005 1.333e+000 15.00
3.000e-003 7.667e+000 86.00
5.800e-003 1.121e+000 88.00
I’m using the TGraph("file.dat", "%lg %lg %*lg") constructor and the plot looks as follows
My end goal is to weigh each point with the 3rd column of the ascii file. I think that using a TH2Poly is the best way, but I’m open to suggestions. The problem is that I’m not sure how to create it. I understand that I need to creat the bins first and then fill them. I think I can create the bins but it’s unclear how to properly fill Any tip is much appreciated!
#include "TH2Poly.h"
#include <vector>
#include <iostream>
#include <fstream>
using namespace std;
void createHistogramFromAsciiFile(const char* filename) {
//Open the file
ifstream file(filename);
//Create vectors
std::vector<double> X, Y, Z;
double x, y, z;
//Fill vectors
while (file >> x >> y >> z) {
X.push_back(x);
Y.push_back(y);
Z.push_back(z);
}
int size = (int) X.size();
//Create a TH2Poly histogram
double x_low = 1.e-5, x_up = 1.e7;
double y_low = 1.e-3, y_up = 1.e3;
double dx = 1.e-3;
double dy = 1.e-3;
TH2Poly *h = new TH2Poly("h", "h", x_low, x_up, y_low, y_up);
// Fill the histogram
for (int i=0; i< size; ++i)
h->AddBin(size, X.data(), Y.data());
//int bins = h->GetBins();
cout << "Total bins = " << h->GetBins() << endl;
//for (int i=0; i<bins; ++i)
//h->Fill(i, Z[i]);
// Draw the histogram
h->Draw("colz");
}