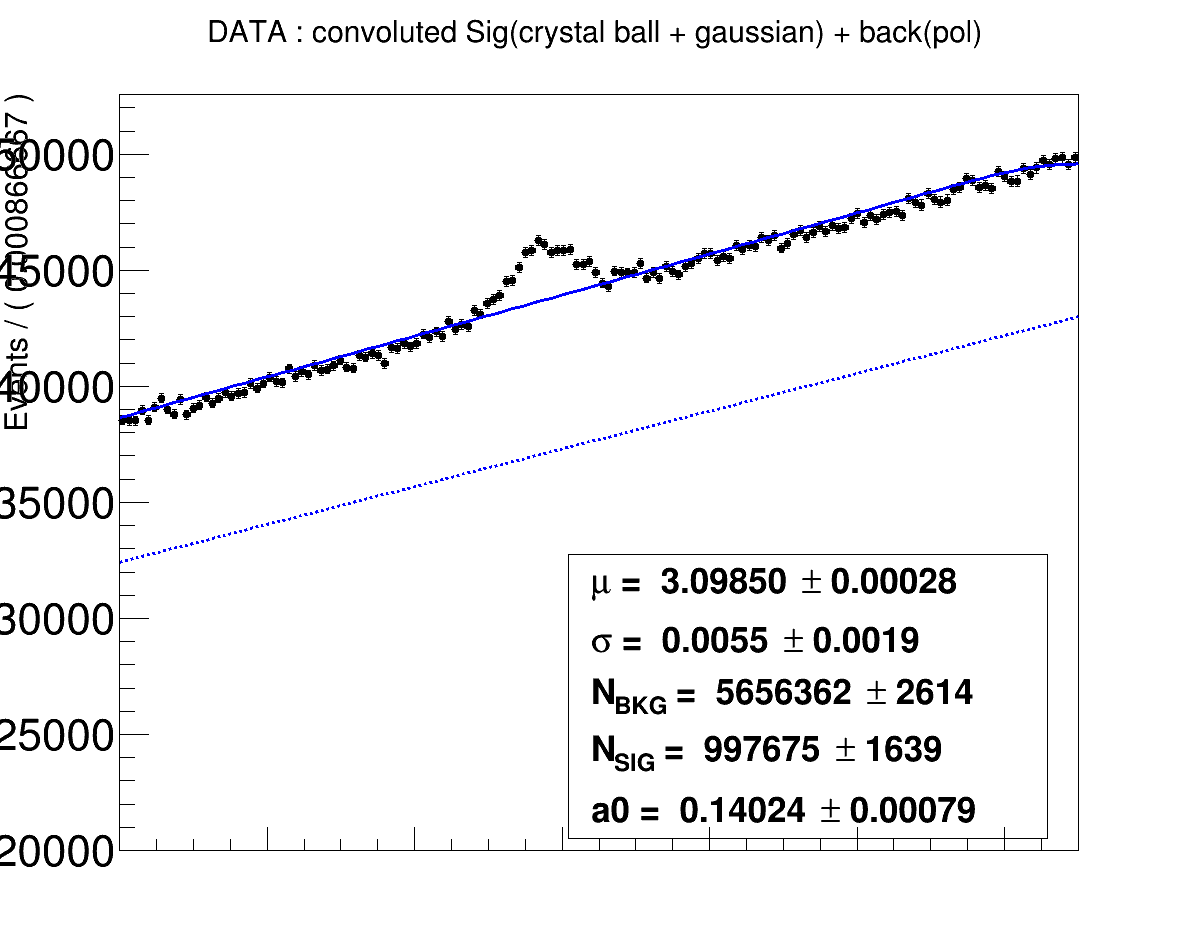
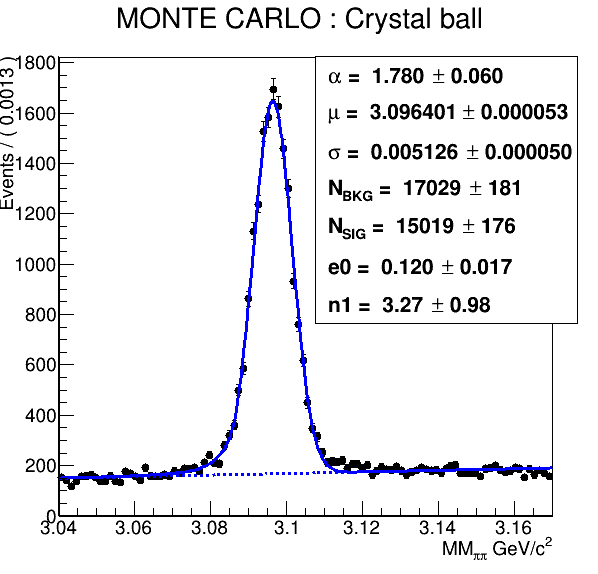
Hello,
I’m trying to fit a histogram with a convoluted (crystal ball + Gaussian ) for the signal part and Chebychev polynomial for the background part.
The idea is to try to fit the signal part in the data with the convoluted Monte carlo shape (crystal ball) and a gaussian.
The fit is failing. you can see the code down below, could you please let me know what I’m doing wrong here?
RooRealVar x("x","MM_{(#pi #pi)} GeV/c^{2}",3.04,3.17) ;
RooDataHist dh("dh","dataset",x,h1);
RooPlot* frame = x.frame(Title("DATA : convoluted Sig(crystal ball + gaussian) + back(pol) ")) ;
dh.plotOn(frame,Name("dh")) ;
//Gaussian
RooRealVar mean1("#mu","mean of gaussians",3.0968,3.0985);
RooRealVar sigma1("#sigma","width of gaussians",0.0025,0.0055);
RooGaussian sig1("sig1", "sig1", x, mean1, sigma1);
//Crystal Ball with fixed parameters from MC
RooRealVar mean2("#mu","mean of gaussians",3.096401);
RooRealVar sigma2("#sigma","width of gaussians",0.005126);
RooRealVar alpha2("#alpha","alpha",1.780);
RooRealVar n2("n2","n",3.27);
RooCBShape sig2("sig2", "sig2", x, mean2, sigma2, alpha2, n2);
//Convolution
RooFFTConvPdf sigg("sigg","gauss",x,sig1,sig2) ;
RooRealVar a0("a0","a0",-2.,2.) ;
//Background
RooChebychev bkg("bkg","background p.d.f.",x,RooArgList(a0));
RooRealVar nsig("N_{SIG}","signal events",0,1000000);
RooRealVar nbkg("N_{BKG}","signal background even0ts",0,100000000);
//Total
RooAddPdf all("all","model",RooArgList(sigg,bkg),RooArgList(nsig,nbkg));
RooFitResult* r = all.fitTo(dh,Extended(kTRUE),Save()) ;
all.paramOn(frame,Layout(0.5,0.90,0.55));
all.plotOn(frame,Components(bkg),LineStyle(kDashed));
all.plotOn(frame,Name("all"));
r->Print();
frame->GetXaxis()->SetTitle("");
frame->GetXaxis()->SetLabelSize(1);
frame->GetYaxis()->SetLabelSize(0.05);
frame->SetMinimum(20000);
frame->Draw();