Hello RoFitters,
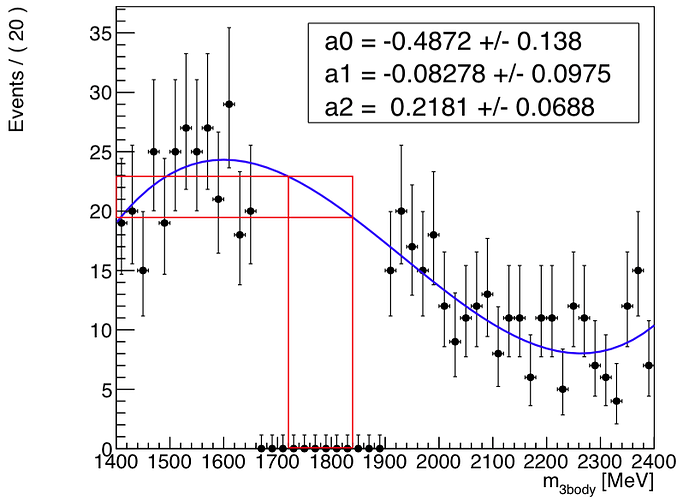
I’m trying to fit the distribution in the attachment in the left and right sidebands together and integrate the resulting function in a small range in the middle of the blinded area (i.e. the signal region…).
The fit function is RooChebychev with 3 parameters.
After fitting successfully the sidebands and plotting everything, I’m calculating the integral in the signal region with createIntegral() but am getting a too high result which I can easily spot if visualising the area below the signal region as you can see in the attached snapshot below.
I guess this is a matter of the normalization of the pdf but I’m clueless on how to work around this.
The brief version of the fit and integral code is
[code]RooRealVar* m3body = new RooRealVar(“m3body”,"m_{3body} [MeV]”,1400,2400);
RooAbsData* UnbinnedDataSet_m3body = new RooDataSet(“data_m3body”,“data_m3body”,RooArgSet(*m3body));
// fill here the dataset with 569 candidates from a tree
m3body->setBins(50); // bin width is 20 MeV
m3body->setRange("range_m3body”, 1400,2400); // full range
m3body->setRange("range_SBleft”, 1400,1660); // left sideband
m3body->setRange(“range_SBright”, 1900,2400); // right sideband
// define the pdf
RooRealVar* a0 = new RooRealVar(“a0”,“a0”, -1.42803e-01, -1.0, 1.0);
RooRealVar* a1 = new RooRealVar(“a1”,“a1”, 2.87161e-02, -1.0, 1.0);
RooRealVar* a2 = new RooRealVar(“a2”,“a2”, -1.45196e-03, -1.0, 1.0);
RooChebychev* bkgm3bodypdf = new RooChebychev(“bkgm3bodypdf”,“bkgm3bodypdf”,*m3body,RooArgSet(*a0,*a1,*a2));
// fit in both sidebands simultaneously
RooFitResult* fitresult_m3body = bkgm3bodypdf->fitTo( *UnbinnedDataSet_m3body,Minos(kTRUE),Range(“range_SBleft,range_SBright”),NormRange(“range_SBleft,range_SBright”),Strategy(2),Save(kTRUE),Timer(kTRUE),NumCPU(2));
// integrate in the signal region (in the heart of the blinded range between the sidebands)
m3body->setRange(“range_m3body_SR”,1718,1838);
RooAbsReal* integralSR0 = bkgm3bodypdf->createIntegral(m3body,Range(“range_m3body_SR”));
RooAbsReal integralSB0left = bkgm3bodypdf->createIntegral(m3body,Range(“range_SBleft”));
RooAbsReal integralSB0right = bkgm3bodypdf->createIntegral(*m3body,Range(“range_SBright”));
float nSB0 = integralSB0left->getVal()+integralSB0right->getVal();
float nSR0 = integralSR0->getVal();
cout << “Events in the side bands=" << nSB0 << endl;
cout << “Events in the signal region=" << nSR0 << endl;[/code]
The numbers I get from the integrals are:
Events in the side bands=710.825 —> should be 569 or very close to it
Events in the signal region =160.302 —> should be around 127 doing the integration "by eye” (see snapshot)
This is basically what I do not understand.
If I do the same with the default TH1F::Fit method in pure ROOT then I get the desirable results (although fitting with Chebychev in ranges is not straightforward), i.e. the integral of the fitted function in the sidebands gives exactly the number of candidates in the sidebands and the integral in the signal region gives the same result that one can extract "by eye”.
Any help is appreciated!
Noam