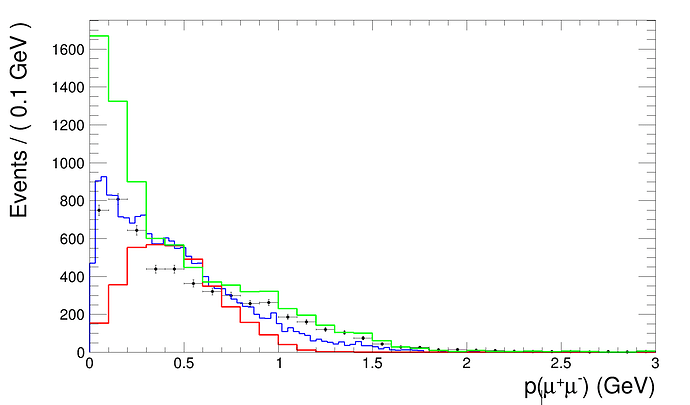
I have a template that’s made out of two datasets combined with RooAddPdf which then are fitted with another dataset as seen in this previous question. My issue is that the resulting histogram has 90 bins, and I would like it to have 30.
I’ve tried adding “RooFit::Binning(30)” to the line
totshape->plotOn(xframe,RooFit::Binning(30));
But that seems to do nothing, the option is completely ignored.
I considered that the bin size of the result maybe has something to do with the bin size of the two datasets used in the template, but those were created using binnedClone() on the the datasets and I didn’t find anything in the documentation regarding an option for the ammount of bins.
Here is the full code:
#include "RooFormulaVar.h"
#include "RooRealVar.h"
#include "RooDataSet.h"
#include "RooDataHist.h"
#include "RooAddPdf.h"
#include "TCanvas.h"
#include "RooPlot.h"
#include "TH1D.h"
void PtFitter()
{
// INPUT FILE
TString flatfile = "DATA_Upsilon_HLTDoubleMu4Acop_Run2011A_invm-8p2-140p0_EXCLSEL_nET-0_ptpair.txt";
TString signal = "Signal1.txt";
TString background = "Background.txt";
// CREATING VARIABLES
RooRealVar* PtPair = new RooRealVar("PtPair","p_{T}(#mu^{+}#mu^{-})",0.0,3.0,"GeV");
RooRealVar* coef1 = new RooRealVar("coef1","PtPair p0",11.2,-150.0,150.0);
RooRealVar* coef2 = new RooRealVar("coef2","PtPair p1",-20.0,-200.0,200.0);
// CREATING DATASETS
RooDataSet *datatmp = 0;
RooDataSet *dataSignal = 0;
RooDataSet *dataBackground = 0;
// READING FILES
datatmp = RooDataSet::read(flatfile,RooArgList(*PtPair));
dataSignal = RooDataSet::read(signal,RooArgList(*PtPair));
dataBackground = RooDataSet::read(background,RooArgList(*PtPair));
// REMOVING BlindedState
RooDataSet * dataSignalReduced = static_cast<RooDataSet*>(dataSignal->reduce(RooFit::SelectVars(*PtPair)));
RooDataSet * dataBackgroundReduced = static_cast<RooDataSet*>(dataBackground->reduce(RooFit::SelectVars(*PtPair)));
// CONVERTING HIST TO PDF
RooDataHist *hist1 = dataSignalReduced->binnedClone();
RooHistPdf histpdf1("histpdf1", "histpdf1", *PtPair, *hist1, 0);
RooDataHist *hist2 = dataBackgroundReduced->binnedClone();
RooHistPdf histpdf2("histpdf2", "histpdf2", *PtPair, *hist2, 0);
// AddPDF
RooAddPdf* totshape = new RooAddPdf("totshape","combined shapes",RooArgList(histpdf1,histpdf2), RooArgList(*coef1), 0);
// FIT
RooFitResult *fitres = totshape->fitTo(*datatmp,RooFit::FitOptions("MHTER"));
// DEFINE HISTOGRAM
TH1D* h1 = new TH1D("h1", "Signal", 30, 0, 3);
TH1D* h2 = new TH1D("h2", "Background", 30, 0, 3);
// FILL HISTOGRAMS WITH DATA
ifstream infile;
infile.open(signal);
double tmp;
while (infile>>tmp){
h1->Fill(tmp);
}
ifstream infile2;
infile2.open(background);
double tmp2;
while (infile2>>tmp2){
h2->Fill(tmp2);
}
h1->SetLineColor(2);
h2->SetLineColor(3);
h1->SetLineWidth(4);
h2->SetLineWidth(4);
// DRAW:
TCanvas *c = new TCanvas("Dilepton signal","Dilepton signal",2000,1200);
RooPlot* xframe = PtPair->frame() ;
datatmp->plotOn(xframe,RooFit::Binning(30));
totshape->plotOn(xframe,RooFit::Binning(30)); //RooFit option doesn't work
xframe->addTH1(h2);
xframe->addTH1(h1);
xframe->Draw();
c->SaveAs("SigPlusBGFit.png");
}