Dear experts,
In my unbinned paramteric analysis, I create PDF’s to model background and fit those to data. I do this by generating various PDF’s like Bernstein polynomials, power law etc and fit them to the entire data i.e without blinding. Later while making a plot of the background fits I blind my signal region using Roofit cutrange.
My first question: Is this the right approach? Or should the data be blinded first and the fitting be performed on blinded data? I will eventually be using the Envelope method for background modeling.
Second question: On using Roofit cutrange
data_RooDataSet.plotOn(frame,RooFit.Binning(tpmass_high-tpmass_low),RooFit.CutRange("unblindreg_1"))
data_RooDataSet.plotOn(frame,RooFit.Binning(tpmass_high-tpmass_low),RooFit.CutRange("unblindreg_2"))
to remove my signal region from the plot, the outputs I get look like this:
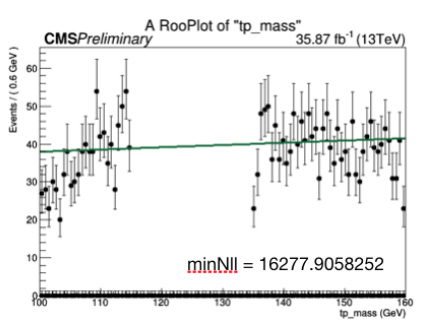
with points at the bottom. Is there a way to avoid these points?
Thanks in advance,
Tanvi