Hi, Lorentz,
Thanks for reply !
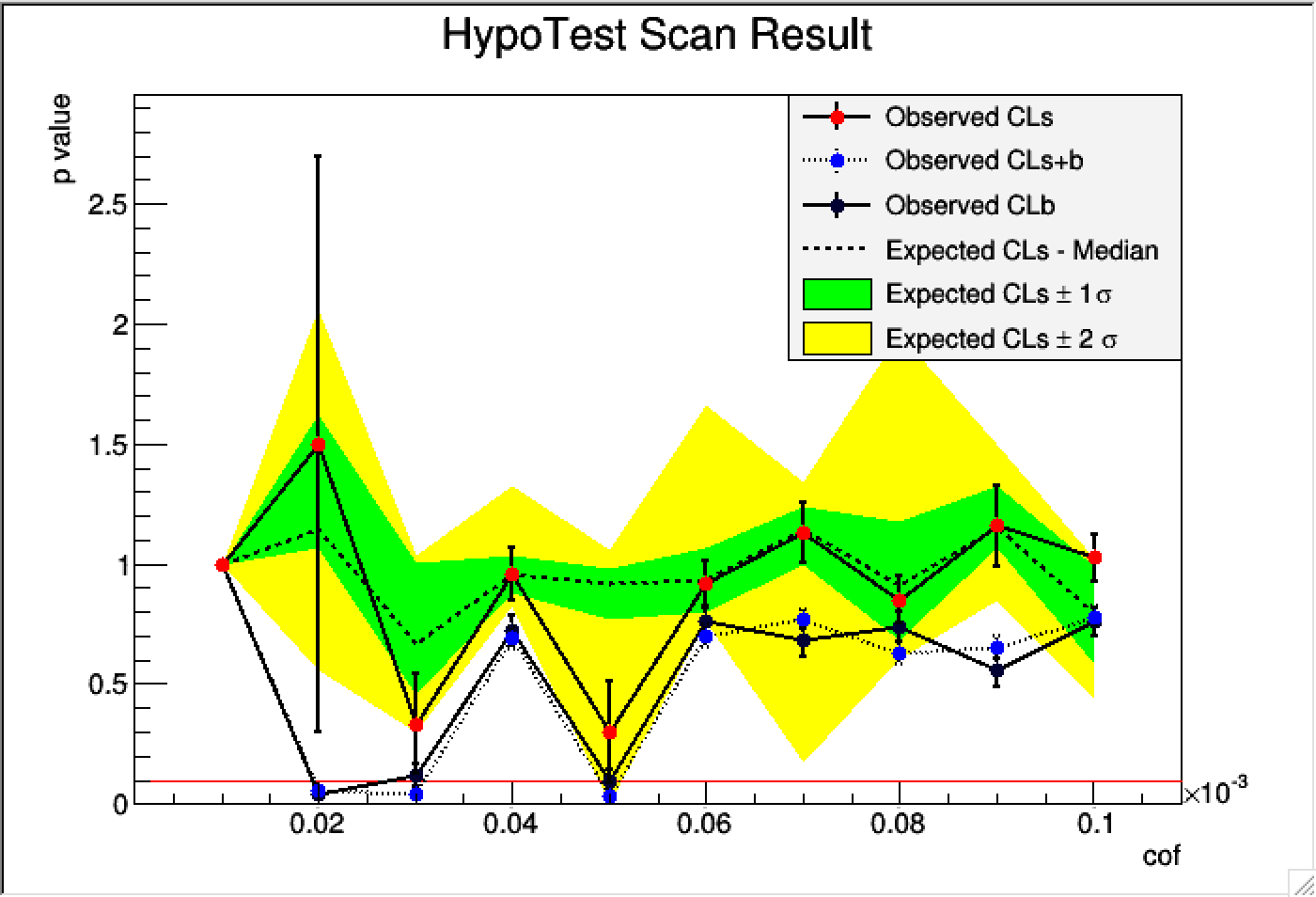
You’re right, the p values of CLb and CLs+b are both very small.
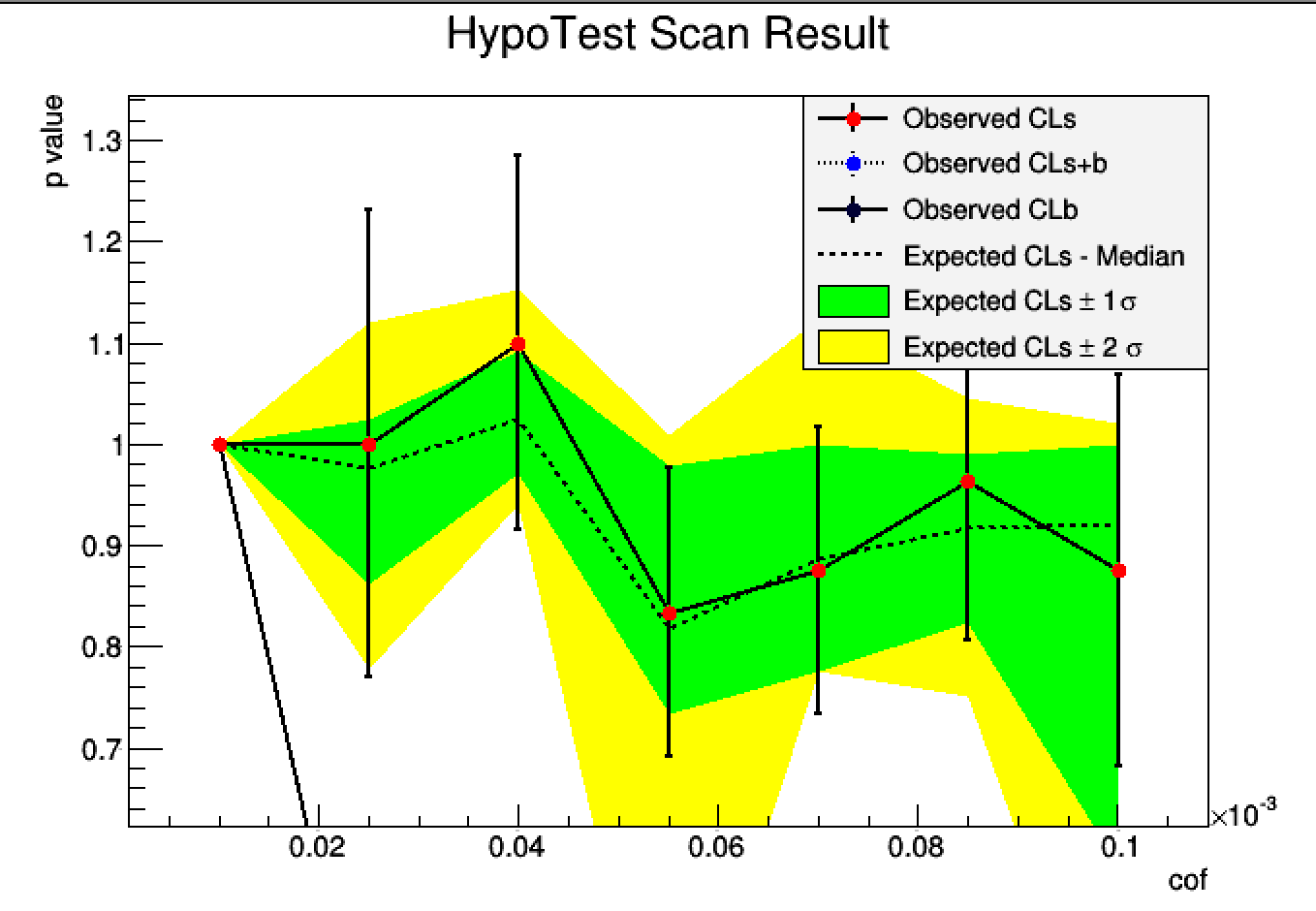
By changing to another model and a smaller dataset, I got a better one.
Comparing to previous plot, this improves significantly I guess.
From this plot, for the cof < 0.06 * 10^-3 = 6*10^-5, the p values of CLb is slightly bigger than CLs+b, so I guess data prefers to CLb(background model only), right ?
Or how to interpret this plot properly ?
Any suggestions for further analysis , for instance, shall I try the analysis of discovery instead ?
The model is attached here, please comment.
BTW, I’ve checked that if POI = 0, the signal model is zero and only the background one survived.
Thanks !
Cheers,
Junhui
#include "TTree.h"
#include "TH1.h"
using namespace RooFit;
using namespace RooStats;
void CountingModel( int nsig = 1,
int nbkg = 1)
{
RooWorkspace w("w");
///~~~~~~~~~~~~~~~~~~ Begining of creating a model p.d.f. ~~~~~~~~~~~~~~///
RooClassFactory::makePdf("BkgLinearPdf","x,A,B") ;
#ifdef __CINT__
gROOT->ProcessLineSync(".L BkgLinearPdf.cxx+") ;
#endif
RooClassFactory::makePdf("OOnePdf","x,cof") ;
#ifdef __CINT__
gROOT->ProcessLineSync(".L OOnePdf.cxx+") ;
#endif
w.factory("BkgLinearPdf:bkg_pdf(x[0.021,10], a[2.5,1.0,30.0], b[-0.015,-0.010])");/// a,interceipt; b, slope.
w.factory("OOnePdf:sig_pdf(x[0.021,10],cof[5.0E-5,1.0E-5,1.0E-4])");
w.factory("SUM:model(nsig[0,20]*sig_pdf, nbkg[0,20]*bkg_pdf)");// for extended model(poisson). 20 from data
///~~~~~~~~~~~~~~~~~~ End of creating a model p.d.f. ~~~~~~~~~~~~~~///
///~~~~~~~ Begining of creating the ModelConfig object and set "b0" as a global observable ~~~///
w.var("nsig")->setVal(nsig); // assign the nsig value defined above into w sapce.
w.var("nbkg")->setVal(nbkg);
w.var("b")->setConstant(true); // needed for being treated as global observables
ModelConfig mc("ModelConfig",&w);/// declare the name and reference of ModelConfig as "mc" and "ModelConfig".
mc.SetPdf(*w.pdf("model"));
mc.SetParametersOfInterest(*w.var("cof"));
mc.SetObservables(*w.var("x"));
w.defineSet("nuisParams","a,nsig,nbkg");
mc.SetNuisanceParameters(*w.set("nuisParams") );
// these are needed for the hypothesis tests
mc.SetSnapshot(*w.var("cof"));
// need now to set the global observable
mc.SetGlobalObservables(*w.var("b"));
mc.Print();
// import model in the workspace
w.import(mc);
///~~~~~~~ End of creating the ModelConfig object and set "b0" as a global observable ~~~///
// import a tree
TFile *treefile = TFile::Open("energyDepositedLess1keV.root");
TTree* tree = (TTree*) treefile->Get("ntuple");
// Define observables x
RooRealVar x("x","x",1,0.021,10) ; // "1" is the initial value of x.
// Import only observables (x)
RooDataSet ds("ds","ds",*w.var("x"),Import(*tree)) ; ///refered from rf102_dataimport.C
ds.Print() ;
ds.add( *w.var("x"));
RooPlot* frame3 = x.frame(Title("Recorded energy in CCDs")) ;
ds.plotOn(frame3) ;
TCanvas* c = new TCanvas("Recorded energy","Recorded energy",800,800) ;
gPad->SetLeftMargin(0.15) ; frame3->GetYaxis()->SetTitleOffset(1.4) ; frame3->Draw() ;
w.import(ds);
w.Print();
TString fileName = "CountingModel.root";
// write workspace in the file (recreate file if already existing)
w.writeToFile(fileName, true);
}