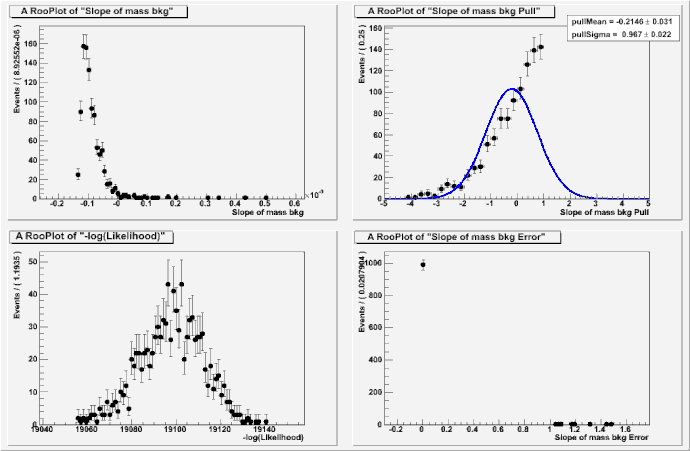
Hi,
Actually, I met this problem some time ago. You know, I just want to use RooPolynomial (1st polynomial) to describe the background, and I use a Gaussian for the signal. I guess this is the very simple usage of RooFit. But there is one thing I could not understand. If I do a toy Monte Carlo with this PDF, the pull of the coefficient would be bad. Please see the attached plot.
I guess that it is cased by the wrong step size? But I failed to find how I could set the step size for the floating variable in RooFit. Could you please give some hints here? It seems to me that setError doesn’t help.
BTW: this is done in Root 5.17/04, but I think the root version doesn’t matter.
Thanks a lot.
Cheers, Jibo
void mass()
{
gSystem->Load("libRooFit");
using namespace RooFit;
RooRealVar *m = new RooRealVar( "m", "invariant mass", 5000, 5600, "MeV/c^{2}");
RooArgSet* observables = new RooArgSet( *m );
/// sig mass
RooRealVar *SigMass = new RooRealVar("SigMass","B mass", 5279, 5000, 5600, "MeV/c^{2}" );
RooRealVar *SigMassRes = new RooRealVar("SigMassRes","B mass resolution", 25, 0., 60, "MeV/c^{2}");
RooAbsPdf *SigMassPdf = new RooGaussian("SigMassPdf","MassPDF of signal",*m,*SigMass,*SigMassRes);
/// bkg mass
RooRealVar *BkgMassSlope = new RooRealVar("BkgMassSlope","Slope of mass bkg", -9.7194e-05, -1.0, 1.0 );
RooAbsPdf *BkgMassPdf = new RooPolynomial("BkgMassPdf","BkgMassPdf",*m, *BkgMassSlope );
RooRealVar *FracSig = new RooRealVar("FracSig","fraction of signal", 0.11, 0, 1.0); //1.2802e-01
RooAbsPdf *MassPdf = new RooAddPdf("MassPdf","mass pdf",
RooArgList(*SigMassPdf,*BkgMassPdf),
RooArgList(*FracSig) );
BkgMassSlope->setError( 1.0e-8 ) ;
RooMCStudy *mgr = new RooMCStudy(*MassPdf, *observables, FitModel(*MassPdf), FitOptions( InitialHesse(true), Minos(false) ) );
mgr->generateAndFit( 1000, 3000 );
TCanvas *c = new TCanvas();
c->Divide(2,2);
c->cd(1);
RooPlot* frame1 = mgr->plotParam(*BkgMassSlope);
frame1->Draw();
c->cd(2);
RooPlot* frame2 = mgr->plotPull(*BkgMassSlope,-5,5,40,kTRUE);
frame2->Draw();
c->cd(3);
mgr->plotNLL()->Draw();
c->cd(4);
mgr->plotError(*BkgMassSlope)->Draw();
}