Hi all,
I am running the following chisquared minimization in Minuit2. I know that as it is, Minuit does not converge, but my question is more about how Minuit explores the values of parameters during the process.
Here is my code:
#include "Minuit2/FunctionMinimum.h"
#include "Minuit2/MnUserParameters.h"
#include "Minuit2/MnUserParameterState.h"
#include "Minuit2/MnPrint.h"
#include "Minuit2/MnMigrad.h"
#include "Minuit2/MnMinos.h"
#include "Minuit2/MnPlot.h"
#include "Minuit2/MnContours.h"
#include "Minuit2/FCNBase.h"
#include <utility> //std::pair
#include <iostream>
#include <iomanip>
#include <string>
#include <vector>
#include <cmath>
#include <Eigen/Dense>
using namespace ROOT::Minuit2;
using namespace Eigen;
const double PI = 4*atan(1);
// Define minimizer class (inherits from FCNBase)
class Minimizer : public FCNBase {
private:
const std::vector< std::vector<double> > &m_data;
// const std::vector<double> &m_mass;
public:
Minimizer(const std::vector< std::vector<double> > &data) : m_data(data) {}
~Minimizer() {}
// functions to find factorial and double factorial of given number - used in th\
e Vqq matrix element
unsigned int doublefactorial(unsigned int number) const
{
if (number == 0 || number == 1)
return 1;
return number*doublefactorial(number-2);
}
unsigned int powerfactorial(unsigned int number) const
{
if (number == 0)
return 2;
return pow(2,number + 1)*powerfactorial(number-1);
}
double diagonalization(double n, double L, double p0, double p1, double p2, double p3) const {
// Declare the number of terms in the expansion and the number of iterations for the numerical integrals
int N1 = 12;
// Declare the variational parameters - values taken from the hybrid decay paper: https://arxiv.org/abs/2005.10850
double alphaInit = 0.005;
// Define the matrices - the dimension can be set to be any of the above indexes since they are generally the same
MatrixXd HMX(N1, N1);
MatrixXd NMX(N1, N1);
MatrixXd KMX(N1, N1);
MatrixXd VMX(N1, N1);
//Declare the variables for the minimum eigenvalue
double minEvalue0 = 0.0; // ground state eigenvalue
double minEvalue1 = 0.0; // first excited state eigenvalue
double minEvalue2 = 0.0; // second excited state eigenvalue
double minEvalue3 = 0.0; // third excited state eigenvalue
std::vector<double> eValues0;
std::vector<double> eValues1;
std::vector<double> eValues2;
std::vector<double> eValues3;
std::vector<double> minEvalues;
// Loop over the variational parameters and the matrix elements (k1=n', l1=m', k2=n, l2=m).
for (double alpha = alphaInit; alpha <= 1.000; alpha += 0.005){
for (int k1 = 1; k1 <= N1; k1++){
for (int k2 = 1; k2 <= N1; k2++){
// Define the full Hamiltonian in configuration space - works for all values of L
// Overlap Matrix
NMX(k1-1,k2-1) = pow(2.0,L + 1)*sqrt(2.0)*pow(k1*k2,0.75 + L/2.0)/(pow(k1+k2,1.5 + L));
// Mass and kinetic terms
KMX(k1-1,k2-1) = (pow(2.0,L + 1)*sqrt(2.0)*pow(k1*k2,0.75 + L/2.0)/(pow(k1+k2,1.5 + L))) * (2.0*p0 + (3.0+2.0*L)*(k1*k2)*alpha*alpha/(p0*(k1+k2)));
// Potential term
VMX(k1-1,k2-1) = (powerfactorial(L)*pow(k1*k2,0.75 + L/2.0)/(doublefactorial(2*L+1)*sqrt(PI)*alpha*pow(k1+k2,2.0+L)))*(2*p1*alpha*alpha*(k1+k2)-3*(L+1)*p2);
}
}
// Diagonalize and print the eigenvalues
HMX = KMX - (4.0/3.0)*VMX;
std::setprecision(8);
GeneralizedSelfAdjointEigenSolver<MatrixXd> es(HMX,NMX);
//pushback eigenvalues into vectors
eValues0.push_back(es.eigenvalues()[0]);
eValues1.push_back(es.eigenvalues()[1]);
eValues2.push_back(es.eigenvalues()[2]);
eValues3.push_back(es.eigenvalues()[3]);
}
// Calculate the minimum eigenvalue and the corresponding parameters
minEvalue0 = eValues0[0];
minEvalue1 = eValues1[0];
minEvalue2 = eValues2[0];
minEvalue3 = eValues3[0];
//cout << "\n";
//cout << "Here is the full list of eigenvalues: " << std::endl;
for(unsigned int i = 0; i < eValues0.size(); i++){
if (eValues0[i] < minEvalue0){
minEvalue0 = eValues0[i];
}
}
for(unsigned int i = 0; i < eValues1.size(); i++){
if (eValues1[i] < minEvalue1){
minEvalue1 = eValues1[i];
}
}
for(unsigned int i = 0; i < eValues2.size(); i++){
if (eValues2[i] < minEvalue2){
minEvalue2 = eValues2[i];
}
}
for(unsigned int i = 0; i < eValues3.size(); i++){
if (eValues3[i] < minEvalue3){
minEvalue3 = eValues3[i];
}
}
//push_back all the minimized eigenvalues into a vector
minEvalues.push_back(minEvalue0);
minEvalues.push_back(minEvalue1);
minEvalues.push_back(minEvalue2);
minEvalues.push_back(minEvalue3);
double eigenvalue = minEvalues[n-1]-p3;//return the desired eigenvalue from minEvalues
return eigenvalue;
}
std::vector<std::string> particleNames ={"eta_b(1S)", "Y(1S)", "chi_b0(1P)", "chi_b1(1P)", "h_b(1P)", "chi_b2(1P)", "Y(2S)", "Y(1D)", "Y(2D)", "chi_b0(2P)", "chi_b1(2P)", "h_b(2P)", "chi_b2(2P)", "Y(3S)", "Y(4S)", "eta_2(2S)"};
double operator() (const std::vector<double> ¶ms) const {
double X2 = 0.0;
double relErr = 0.0;
for (int i=0; i< m_data[0].size(); i++) {
double n = m_data[0][i];
double L = m_data[1][i];
double f = diagonalization(n, L, params[0], params[1], params[2],params[3]);
relErr = abs(m_data[2][i]-f)*100.0/m_data[2][i];
X2 += (m_data[2][i] - f)*(m_data[2][i] - f);
std::cout << "params: " << params[0] << ", " << params[1] << ", "
<< params[2] << ", " << params[3] << std::endl;
std::cout << std::setw(10) << particleNames[i] << ", exp[" << i << "] = " << std::setw(10) << m_data[2][i]
<< " || theory[" << i << "] = " << std::setw(10) << f << " || relative error = " << relErr << " %" << std::endl;
}
std::cout << "\n";
std::cout << "params: " << params[0] << ", " << params[1] << ", " << params[2] << ", " << params[3] << std::endl;
std::cout << X2 << std::endl;
return X2;
}
//set to 1 for chi-sq and 0.5 for log-likelihood
double Up() const {return 1.0;}
};
int main(){
std::vector< std::vector<double> > data(3);
// bottomonium experimental states
//eta (1S) n=1, L=0; N=0
data[0].push_back(1.0);
data[1].push_back(0.0);
data[2].push_back(9.390);
//Y (1S) n=1, L=0; N=0
data[0].push_back(1.0);
data[1].push_back(0.0);
data[2].push_back(9.4603);
//chi_b0 (1P) n=2, L=1; N=0
data[0].push_back(1.0);
data[1].push_back(1.0);
data[2].push_back(9.8594);
//chi_b1 (1P) n=2, L=1; N=0
data[0].push_back(1.0);
data[1].push_back(1.0);
data[2].push_back(9.8928);
//h_b (1P) n=2, L=1; N=0
data[0].push_back(1.0);
data[1].push_back(1.0);
data[2].push_back(9.899);
//chi_b2 (1P) n=2, L=1; N=0
data[0].push_back(1.0);
data[1].push_back(1.0);
data[2].push_back(9.9122);
//Y (2S) n=2, L=0; N=1
data[0].push_back(2.0);
data[1].push_back(0.0);
data[2].push_back(10.0233);
//Y (1D) n=3, L=2; N=0
data[0].push_back(1.0);
data[1].push_back(2.0);
data[2].push_back(10.1611);
//Y2 (1D) n=3, L=2; N=0
data[0].push_back(1.0);
data[1].push_back(2.0);
data[2].push_back(10.1637);
//chi_b0 (2P) n=3, L=1; N=1
data[0].push_back(2.0);
data[1].push_back(1.0);
data[2].push_back(10.2325);
//chi_b1 (2P) n=3, L=1; N=1
data[0].push_back(2.0);
data[1].push_back(1.0);
data[2].push_back(10.2555);
//h_b (2P) n=3, L=1; N=1
data[0].push_back(2.0);
data[1].push_back(1.0);
data[2].push_back(10.2598);
//chi_b2 (2P) n=3, L=1; N=1
data[0].push_back(2.0);
data[1].push_back(1.0);
data[2].push_back(10.2686);
//Y (3S) n=3, L=0; N=2
data[0].push_back(3.0);
data[1].push_back(0.0);
data[2].push_back(10.3552);
//Y (4S) n=4, L=0; N=3
data[0].push_back(4.0);
data[1].push_back(0.0);
data[2].push_back(10.580);
//Y (2S) n=4, L=1, N=2
data[0].push_back(2.0);
data[1].push_back(0.0);
data[2].push_back(9.999);
//Create FCN function
Minimizer fcn(data);
//Create parameters
double mq = 4.7;
double alphaS = 0.34;
double b = 0.19;
double shift = 0.1;
MnUserParameters uParams;//use params[i]/5 error
uParams.Add("mq", mq, 0.2*mq);
uParams.Add("alphaS", alphaS, 0.2*alphaS);
uParams.Add("b", b, 0.2*b);
uParams.Add("shift", shift, 0.2*shift);
std::cout << "Computing the values of the parameters..." << std::endl;
for (int i=0;i<data[0].size();i++) {
std::cout << i << " " << data[2][i] << " : " << fcn.diagonalization(data[0][i],data[1][i],mq,alphaS,b) << std::endl;
}
//minimize
//Minimizer fcn(data);
//Set strategy to High
MnStrategy strategy;
strategy.SetHighStrategy();
MnMigrad migrad(fcn, uParams);
//fix a parameter
//migrad.Fix("shift");
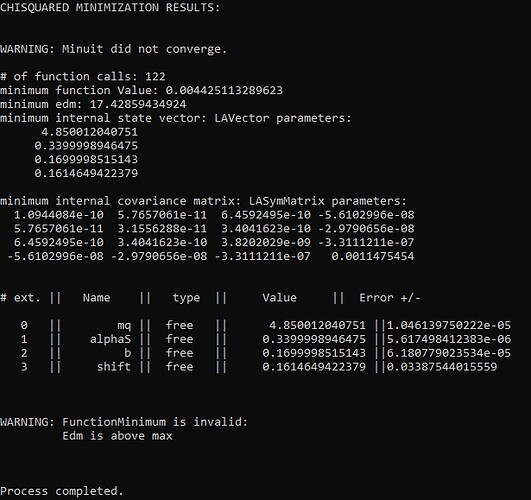
FunctionMinimum min = migrad();
std::cout << "\n";
std::cout << "CHISQUARED MINIMIZATION RESULTS:" << std::endl;
std::cout << "\n";
std::cout << min << std::endl;
std::cout << "Process completed." << std::endl;
return 0;
}
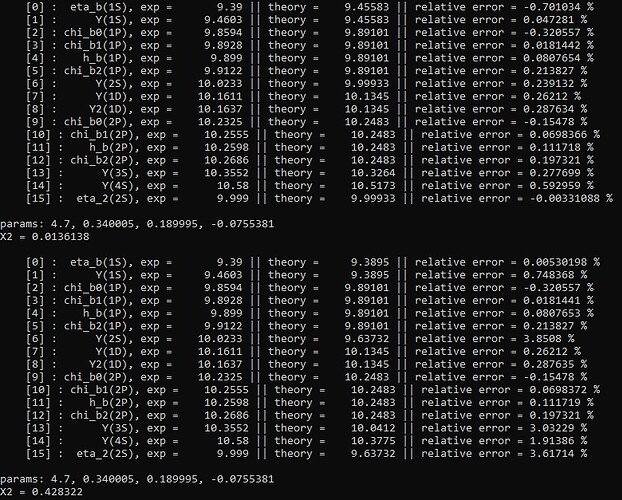
It is my understanding that in order to find the minimum for the objective function, Minuit2 will ‘explore’ various values of the parameters. But when I echo print the values of the parameters to the screen this is what I get.
So, the values of the parameters are the same in both runs, but the X2 value AND the theory masses produced by my code with those parameters are different???
Am I missing something in the way Minuit2 works? Any help would be greatly appreciated. Thanks.
Christian
ROOT Version: Minuit2 StandAlone version 6.15
Platform: Linux Mint 17
Compiler: gcc