Hello,
I have a problem with a root macro. What I basically want to do is that I have one big root tree where several single plots/histograms of length 900000 each are after each other. Then I want to fit a standard gaussian distribution to each single plot what I do via a loop and extract the fitting parameters. Unfortunately, root seems to remember the previous fits, because the first fit works and then I get problems. This always happens no matter at which positon/ with which of the single histograms I start, so it must have someting to do with the fitting in the loop. I do delete the histogram and the function at the end, but despite that, it happens again and again.
My full code is:
void get_baseline_cuts_gaus(){
TFile *file1 = TFile::Open(“datatree.root”);
TTree treeInput = (TTree) file1->Get(“treeData”);
unsigned int evt_n;
double peak_integral;
double peak_maximum;
double time_maximum;
double baseline_mean;
double baseline_rms;
treeInput->SetBranchAddress("evt_n", &evt_n);
treeInput->SetBranchStatus("evt_n",0);
treeInput->SetBranchAddress("peak_integral",&peak_integral);
treeInput->SetBranchStatus("peak_integral",1);
treeInput->SetBranchAddress("peak_maximum",&peak_maximum);
treeInput->SetBranchStatus("peak_maximum",0);
treeInput->SetBranchAddress("time_maximum",&time_maximum);
treeInput->SetBranchStatus("time_maximum",0);
treeInput->SetBranchAddress("baseline_mean",&baseline_mean);
treeInput->SetBranchStatus("baseline_mean",0);
treeInput->SetBranchAddress("baseline_rms",&baseline_rms);
treeInput->SetBranchStatus("baseline_rms",1);
string runnumber[15] = {"Run00695", "Run00696", "Run00697", "Run00698", "Run00699", "Run00700", "Run00701", "Run00702", "Run00703", "Run00704", "Run00705", "Run00706", "Run00707", "Run00708", "Run00709"};
double maximum[15] = {0};
double baseline_cut_gaus[15][4] = {0};
int number = 0;
double cut[4][4] = {0};
for ( int j = 0; j < 2; j++ ){
//~ int j = 1;
number += 900000;
//~ number += 900000;
//~ number += 900000;
//~ number += 900000;
TH1F *hist1 = new TH1F ("hist", "hist", 400, 0, 1 );
for( int i = number; i < ( 900000 + number ); i++){
treeInput->GetEntry(i);
hist1->Fill(baseline_rms);
}
number += 900000;
cout << "number:\t" << number << endl;
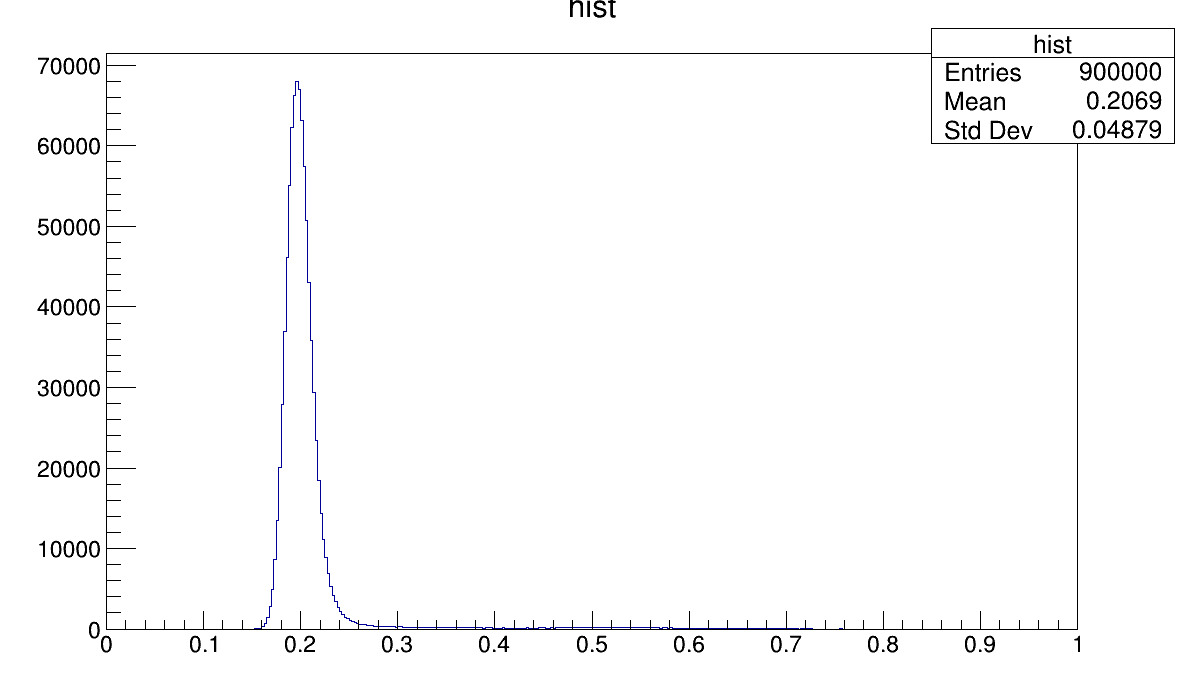
int binmax = hist1->GetMaximumBin();
maximum[j] = hist1->GetXaxis()->GetBinCenter(binmax);
cout << "maximum:\t" << maximum[j] << endl;
cout << "Binmax:\t:" << binmax <<endl;
// fit gaus to baseline:
double left_end = maximum[j] - 0.02;
double right_end = maximum[j] + 0.015;
TF1 *g1 = new TF1("g1", "gaus", left_end, right_end);
hist1->Fit(g1, "R") ;
hist1->Draw();
g1->Draw("same");
double amplitude = g1->GetParameter(0);
double mean = g1->GetParameter(1);
double sigma = g1->GetParameter(2);
cout << "amplitude:\t" << amplitude << endl;
cout << "mean:\t" << mean << endl;
cout << "sigma:\t" << sigma << endl;
for ( int n = 0; n < 4; n++){
//~ int n = 1;
baseline_cut_gaus[j][n-1] = mean + n * sigma;
cout << "baseline_cut_gaus:\t" << baseline_cut_gaus[j][n-1] << endl;
}
delete g1;
delete hist1;
}
}
And I get the following error:
FCN=435.937 FROM MIGRAD STATUS=CONVERGED 74 CALLS 75 TOTAL
EDM=2.31573e-10 STRATEGY= 1 ERROR MATRIX UNCERTAINTY 2.1 per cent
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 Constant 6.88087e+04 1.08427e+02 -7.18223e-03 2.16701e-07
2 Mean 1.97145e-01 2.06284e-05 -7.33589e-09 -3.76929e-02
3 Sigma 1.18415e-02 2.22333e-05 -6.04209e-07 5.25611e-02
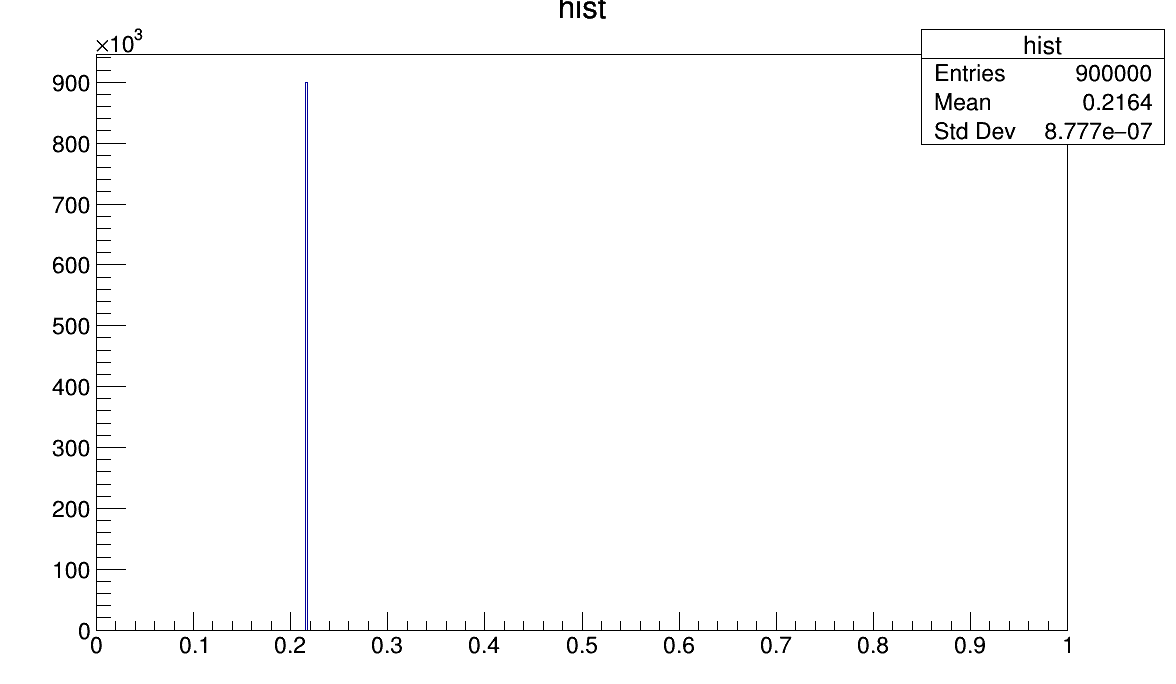
FCN=8.84646e-13 FROM HESSE STATUS=NOT POSDEF 16 CALLS 97 TOTAL
EDM=1.76722e-12 STRATEGY= 1 ERR MATRIX NOT POS-DEF
EXT PARAMETER APPROXIMATE STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 Constant 1.16370e+06 1.82904e+04 2.77448e-01 1.53354e-09
2 Mean 3.94067e-01 5.43747e-03 9.39529e-08 -5.15770e-03
3 Sigma 2.48038e-01 7.58441e-03 2.21709e-07 2.76288e-03
So the first fit converges, but the second not.
Has anyone an idea why that is and how to solve this problem?
Kind regards
Judith