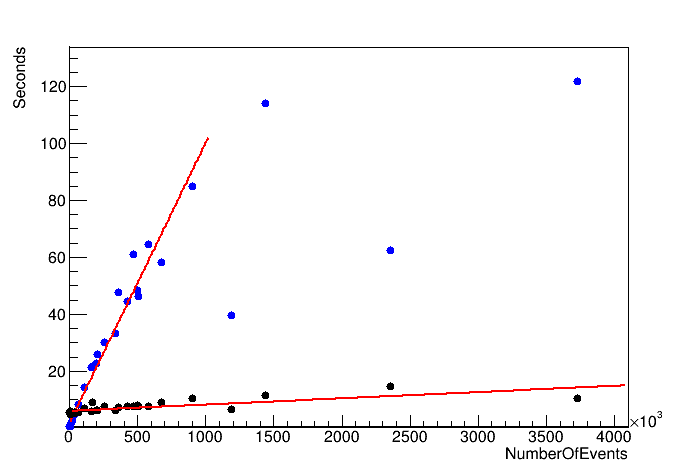
Some of your measurements are surprising, e.g. python @ 2353543 is way lower than what a fit would suggest.
For python I get 2s + 1E-4s*N, for RDF it’s 6s + 2E-6s*N. The constant time isn’t what it should be - and we’ll improve likely both parameters by factors during the next 18 months: we know exactly what we want to do to fix them. (FYI: bulk processing of RNTuple)!