Hi! So, I read ROOT primer and about to complete ROOT manual and ,I have 2 questions about dataset provided in the form of ROOT files.
Firstly, given a root file in open-data, how to carry out analysis with it ? Is there any tutorial or some course that took place in the past?
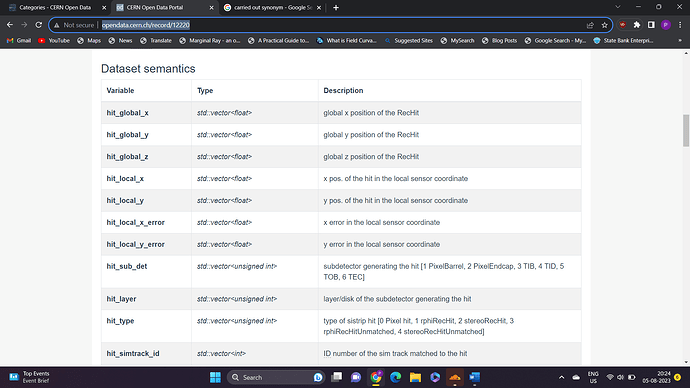
Secondly, where to find information about the dataset semantics provided …as the one given in the site along with dataset does not seems so clear ?
I’m attaching the link for the dataset that I am currently want to work on and , the image of some of the dataset semantics to clear what I am asking.
To see what’s inside a ROOT file, you can use rootls in a terminal, e.g.:
rootls -l hsimple.root
TProfile Apr 12 17:27 2022 hprof;1 "Profile of pz versus px"
TH1F Apr 12 17:27 2022 hpx;1 "This is the px distribution"
TH2F Apr 12 17:27 2022 hpxpy;1 "py vs px"
TNtuple Apr 12 17:27 2022 ntuple;1 "Demo ntuple"
(that’s a small “L” in rootls -l ...; for more options, just do rootls -h).
The first column is the type of object. After the timestamps is the name (what you’ll use to ‘get’ the object from the file) and at the end is the description, but note that descriptions depend on whoever created the file, so they may or may not be useful (or might even be empty).
You get more details with the option ‘-t’
rootls -t hsimple.root
TProfile Apr 12 17:27 2022 hprof;1 "Profile of pz versus px"
TH1F Apr 12 17:27 2022 hpx;1 "This is the px distribution"
TH2F Apr 12 17:27 2022 hpxpy;1 "py vs px"
TNtuple Apr 12 17:27 2022 ntuple;1 "Demo ntuple"
px "px" 100284
py "py" 100284
pz "pz" 100284
random "random" 100300
i "i" 100280
Cluster INCLUSIVE ranges:
- # 0: [0, 24999]
The total number of clusters is 1
A TNtuple is similar to a TTree (which is whay you’ll see more often), and you can read it like reading a TTree. If you see the above TNtuple as a TTree, the branches are px, py, … To see the type of each one, you can first open the ROOT file in ROOT and then do nameoftree->Print(), e.g.:
root hsimple.root
------------------------------------------------------------------
| Welcome to ROOT 6.26/10 https://root.cern |
| (c) 1995-2021, The ROOT Team; conception: R. Brun, F. Rademakers |
| Built for linuxx8664gcc on Nov 16 2022, 10:42:54 |
| From tags/v6-26-10@v6-26-10 |
| With c++ (Ubuntu 11.3.0-1ubuntu1~22.04) 11.3.0 |
| Try '.help', '.demo', '.license', '.credits', '.quit'/'.q' |
------------------------------------------------------------------
root [0]
Attaching file hsimple.root as _file0...
(TFile *) 0x55e3c4052740
root [1] ntuple->Print()
******************************************************************************
*Tree :ntuple : Demo ntuple *
*Entries : 25000 : Total = 504176 bytes File Size = 429478 *
* : : Tree compression factor = 1.17 *
******************************************************************************
*Br 0 :px : Float_t *
*Entries : 25000 : Total Size= 100755 bytes File Size = 92913 *
*Baskets : 4 : Basket Size= 32000 bytes Compression= 1.08 *
*............................................................................*
*Br 1 :py : Float_t *
*Entries : 25000 : Total Size= 100755 bytes File Size = 92934 *
*Baskets : 4 : Basket Size= 32000 bytes Compression= 1.08 *
*............................................................................*
*Br 2 :pz : Float_t *
*Entries : 25000 : Total Size= 100755 bytes File Size = 91194 *
*Baskets : 4 : Basket Size= 32000 bytes Compression= 1.10 *
*............................................................................*
*Br 3 :random : Float_t *
*Entries : 25000 : Total Size= 100787 bytes File Size = 90003 *
*Baskets : 4 : Basket Size= 32000 bytes Compression= 1.11 *
*............................................................................*
*Br 4 :i : Float_t *
*Entries : 25000 : Total Size= 100747 bytes File Size = 61673 *
*Baskets : 4 : Basket Size= 32000 bytes Compression= 1.63 *
*............................................................................*
root [2]
so px is a Float_t, and so on.
As for how to get/see the data and analyse it, it depends on what object it is, and is explained in the Primer, Manual and Tutorials (check out the links in Get Started).
Hi,
This is because there is no ntuple object in your file. Check the output of
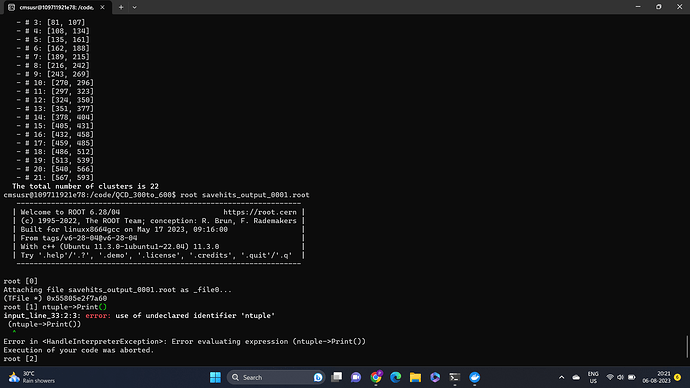
rootls -l root://eospublic.cern.ch//eos/opendata/cms/datascience/TrackerRecHitProducerTool/QCD300to600_RunI_8TeV/step3_QCD300to600/savehits_output_0001.root
once again. There is only a TTree called hits_tree, for which you can do hits_tree->Print().
Thankyou so much. It worked! ![]()
I got one more doubt regarding the output we receive from rootls -t command that at the end we have clusters. Can you please share some link about clusters and what do these inclusive ranges mean?