Hi Wouter,
many thanks for your reply !!
I’ve corrected there error and added the “fitRange” as arguments
to my extended pdf, i see two effects:
- output with a lot of lines like:
RooAbsRealLValue::inFitRange(m): value 115.625 rounded down to max limit 110
RooAbsRealLValue::inFitRange(m): value 116.875 rounded down to max limit 110
RooAbsRealLValue::inFitRange(m): value 118.125 rounded down to max limit 110
RooAbsRealLValue::inFitRange(m): value 119.375 rounded down to max limit 110
RooAbsRealLValue::inFitRange(m): value 40 rounded up to min limit 70
RooAbsRealLValue::inFitRange(m): value 120 rounded down to max limit 110
RooAbsRealLValue::inFitRange(m): value 60 rounded up to min limit 70
RooAbsRealLValue::inFitRange(m): value 50 rounded up to min limit 70
RooAbsRealLValue::inFitRange(m): value 45 rounded up to min limit 70
i suppose for every points outside my fitRange, is it normal ?
2) a segmentation violation (ooops) :
[code]RooAddPdf::syncCoefProjList(model) creating coefficient projection integrals
from current normalization: (m)
with current range:
to reference normalization: (m)
*** Break *** segmentation violation
Generating stack trace…
0x00002aaaaf46ac95 in RooAddPdf::syncCoefProjList(RooArgSet const*, RooArgSet const*, char const*) const + 0x825 from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf46b852 in RooAddPdf::evaluate() const + 0x32 from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf44f7fa in RooAbsPdf::getVal(RooArgSet const*) const + 0xca from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf515216 in RooRealIntegral::evaluate() const + 0x3a6 from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf450bff in RooAbsReal::traceEval(RooArgSet const*) const + 0x1f from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf450ded in RooAbsReal::getVal(RooArgSet const*) const + 0x12d from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf769a59 in RooScaledFunc::operator()(double const*) const + 0x35 from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf494a8a in RooCurve::addPoints(RooAbsFunc const&, double, double, int, double, double, RooCurve::WingMode) + 0x19a from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf495457 in RooCurve::RooCurve(RooAbsReal const&, RooAbsRealLValue&, double, double, int, double, RooArgSet const*, double, double, bool, RooCurve::WingMode) + 0x3b7 from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf45350f in RooAbsReal::plotOn(RooPlot*, RooAbsReal::PlotOpt) const + 0xa1f from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf455d72 in RooAbsReal::plotOn(RooPlot*, RooLinkedList&) const + 0xaf2 from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf44c999 in RooAbsPdf::plotOn(RooPlot*, RooLinkedList&) const + 0x4f9 from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf44fff7 in RooAbsReal::plotOn(RooPlot*, RooCmdArg const&, RooCmdArg const&, RooCmdArg const&, RooCmdArg const&, RooCmdArg const&, RooCmdArg const&, RooCmdArg const&, RooCmdArg const&, RooCmdArg const&, RooCmdArg const&) const + 0x197 from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaaf57ea8e in from /home/bellomo/software/root/lib/libRooFit.so
0x00002aaaab60e73f in G__call_cppfunc + 0x24f from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab5fad11 in G__interpret_func + 0x17b1 from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab5efe6c in G__getfunction + 0x201c from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab67a1fe in G__getstructmem + 0x62e from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab671dd8 in G__getvariable + 0xb98 from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab5cbfcb in G__getitem + 0xab from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab5cfe18 in G__getexpr + 0x2af8 from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab627921 in G__exec_function + 0x91 from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab62cc89 in G__exec_statement + 0x30e9 from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab5bb610 in G__exec_tempfile_core + 0x300 from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab5bb8eb in G__exec_tempfile + 0xb from /home/bellomo/software/root/lib/libCint.so
0x00002aaaab639d53 in G__process_cmd + 0x65e3 from /home/bellomo/software/root/lib/libCint.so
0x00002aaaaae26480 in TCint::ProcessLine(char const*, TInterpreter::EErrorCode*) + 0x230 from /home/bellomo/software/root/lib/libCore.so
0x00002aaaaad6e295 in TApplication::ProcessFile(char const*, int*) + 0x885 from /home/bellomo/software/root/lib/libCore.so
0x00002aaaaad6d542 in TApplication::ProcessLine(char const*, bool, int*) + 0x4c2 from /home/bellomo/software/root/lib/libCore.so
0x00002aaaaca31524 in TRint::HandleTermInput() + 0x1b4 from /home/bellomo/software/root/lib/libRint.so
0x00002aaaaca30167 in TTermInputHandler::Notify() + 0x17 from /home/bellomo/software/root/lib/libRint.so
0x00002aaaaca31b8d in TTermInputHandler::ReadNotify() + 0xd from /home/bellomo/software/root/lib/libRint.so
0x00002aaaaaeaf213 in TUnixSystem::CheckDescriptors() + 0x1b3 from /home/bellomo/software/root/lib/libCore.so
0x00002aaaaaeb3119 in TUnixSystem::DispatchOneEvent(bool) + 0x5c9 from /home/bellomo/software/root/lib/libCore.so
0x00002aaaaade3c45 in TSystem::InnerLoop() + 0x15 from /home/bellomo/software/root/lib/libCore.so
0x00002aaaaade3bdd in TSystem::Run() + 0x6d from /home/bellomo/software/root/lib/libCore.so
0x00002aaaaad6d6cf in TApplication::Run(bool) + 0x1f from /home/bellomo/software/root/lib/libCore.so
0x00002aaaaca30584 in TRint::Run(bool) + 0x2c4 from /home/bellomo/software/root/lib/libRint.so
0x000000000040104d in main + 0x4d from /home/bellomo/software/root/bin/root.exe
0x00002aaaad2a9d00 in __libc_start_main + 0xb0 from /lib/libc.so.6
0x0000000000400f6a in TApplicationImp::ShowMembers(TMemberInspector&, char*) + 0x3a from /home/bellomo/software/root/bin/root.exe[/code]
if i replace in my code (reported at the end) this line:
RooAddPdf model("model","signal+bg",RooArgList(signalE,backE));
with
RooAddPdf model("model","signal+bg",RooArgList(signalE,backE),RooArgList(*Ns,*Nb));
the program ends without segmentation violation.
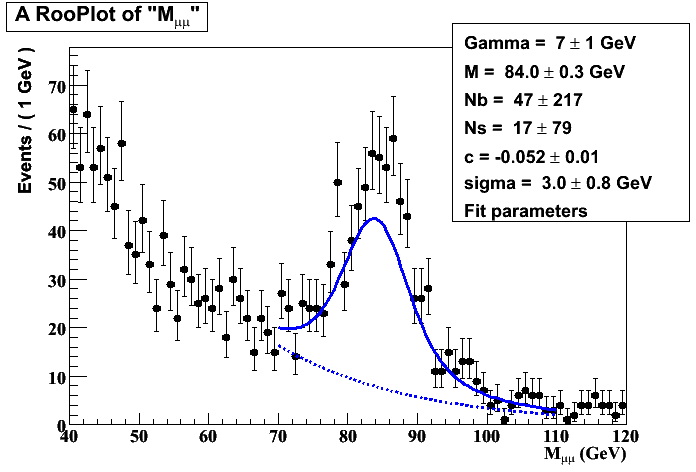
i’ve attached the resulting plot, as you can see the values of Ns, Nb reported don’t seem to be correct and erros are too bigger.
Moreover Ns must be greater that Nb.
There must be some error but i can’t see where !!
Thanks in advance for any comment.
This is my code:
[code]RooRealVar* m = new RooRealVar(“m”,“M_{#mu#mu}”,40,120,“GeV”);
m->setRange(“fitRange”,70,110);
RooRealVar* M = new RooRealVar(“M”,“Z mass”,90.,70.,110.,“GeV”);
RooRealVar* sigma = new RooRealVar(“sigma”,“resolution”,2.,0.1,30.,“GeV”);
RooRealVar* Gamma = new RooRealVar(“Gamma”,“Z decay rate”,2.,0.1.,30.,“GeV”);
RooRealVar* c = new RooRealVar(“c”,“costant”,-0.1,0.);
RooRealVar* Ns = new RooRealVar(“Ns”,“Number of signal events”,100,0.,5000.);
RooRealVar* Nb = new RooRealVar(“Nb”,“Number of background events”,100,0.,5000.);
// signal
RooVoigtian signal(“signal”,“Breit Wigner plus Resol Model”,*m,*M,*Gamma,*sigma);
RooExtendPdf signalE(“signalE”,“Breit Wigner plus Resol (ext)”,signal,*Ns,“fitRange”);
// background
RooExponential back(“back”,“Exponential background Model”,*m,*c);
RooExtendPdf backE(“backE”,“Exponential background Model (ext)”,back,*Nb,“fitRange”);
// model
RooAddPdf model(“model”,“signal+bg”,RooArgList(signalE,backE));
// get data from histos
RooDataHist* data = new RooDataHist(“data”,“data”,RooArgList(*m),histo);
RooFitResult *fitres = model.fitTo(*data,Range(“fitRange”),Extended(),Minos(kFALSE),Save(kTRUE));
RooPlot* frame1=m->frame();
data.plotOn(frame1);
model.plotOn(frame1,Range(“fitRange”));
model.plotOn(frame1,Components(backE),Range(“fitRange”),LineStyle(kDashed));
model.paramOn(frame1,data,“Fit parameters”,1,“NELU”);
frame1->Draw();[/code]
Bye,
Max.