Naive question about reading DAOD_PHYS into RDataFrame.
So I’d like to read information about jets from a DAOD_PHYS file (one of these, for a concrete example; Cern Authentication). DAOD_PHYSLITE files are easy, I can access the column “AnalysisJetsAuxDyn.pt” to get jet pt. For example;
import ROOT; ROOT.xAOD.Init()
r_frame = ROOT.RDataFrame("CollectionTree", "/path/to/PHYSLITE.root")
filtered = r_frame.Filter("AnalysisJetsAuxDyn.pt.size()>0")
print(filtered.Count().GetValue())
And this prints the number of events with at least one jet. Equally, I could access the indices of “AnalysisJetsAuxDyn.pt” to use the jet’s pt to make filter/new columns, etc.
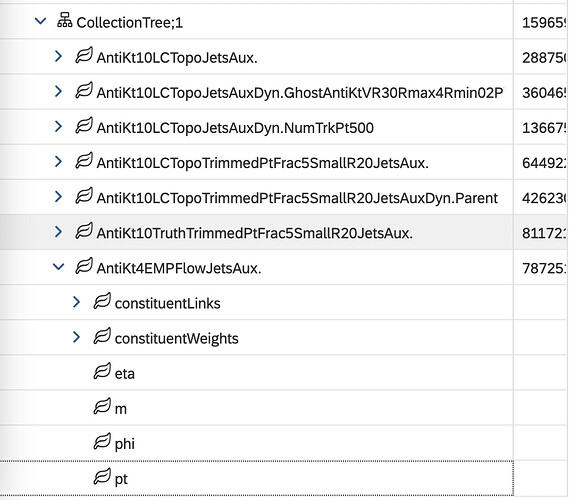
But in the PHYS files, I can see a branch “AntiKt4EMPFlowJetsAux” which has a “pt” leaf that looks great. It plots fine in a TBrowser too. I cannot figure out how to read it in RDataFrame though;
I tried “AntiKt4EMPFlowJetsAux.pt”, but apparently that isn’t what’s needed;
input_line_220:2:35: error: use of undeclared identifier 'AntiKt4EMPFlowJetsAux'
auto lambda0 = [](){return argmax(AntiKt4EMPFlowJetsAux.pt)
Is there a way to access these variables?
Please read tips for efficient and successful posting and posting code
ROOT Version: 6.24/06
Platform: NAME=“CentOS Linux” VERSION=“7 (Core)” ID_LIKE=“rhel fedora” CLUSTER=“sunrise”
Compiler: using pyroot