Dear RDataFrame experts.
I’m trying to run RDF distributed on Swan-Spark.
Swan initialised with /cvmfs/sft.cern.ch/lcg/views/LCG_105a_swan
I have two questions.
- how I pass the voms-provy to RDF
- how I pass my user library .h with basic functions to RDF.
below more details of the problem encountered
Thanks
Maria
=======================================================================
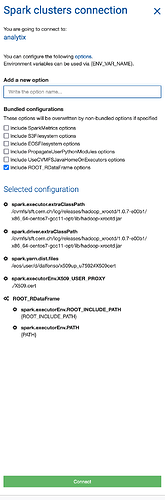
Regarding 1) I initialise spark like this (after checking with swan experts)
.
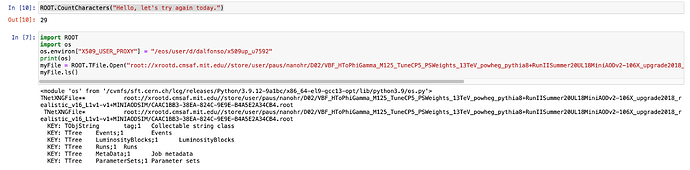
I can read remote file from a cell notebook but I encounter this error when I read with RDF.
File "/cvmfs/sft.cern.ch/lcg/views/LCG_105a_swan/x86_64-el9-gcc13-opt/lib/ROOT/_pythonization/_tfile.py", line 103, in _TFileOpen
raise OSError('Failed to open file {}'.format(str(args[0])))
OSError: Failed to open file root://xrootd.cmsaf.mit.edu//store/user/paus/nanohr/D02/VBF_HToPhiGamma_M125_TuneCP5_PSWeights_13TeV_powheg_pythia8+RunIISummer20UL18MiniAODv2-106X_upgrade2018_realistic_v16_L1v1-v1+MINIAODSIM/CAAC1BB3-38EA-824C-9E9E-B4A5E2A34CB4.root
note works when reading a dataset in my /eos/user/d/dalfonso/ …
=======================================================================
Regarding 2)
I initialise RDF in this way
def init():
ROOT.gInterpreter.ProcessLine('#include "/eos/home-d/dalfonso/SWAN_projects/Hrare/JULY_exp/myLibrary.h"')
import pyspark
RDataFrame = ROOT.RDF.Experimental.Distributed.Spark.RDataFrame # for CERN
RunGraphs = ROOT.RDF.Experimental.Distributed.RunGraphs
ROOT.RDF.Experimental.Distributed.initialize(init)
dfINI = RDataFrame("Events", files, sparkcontext=sc, npartitions=NPARTITIONS) # at CERN
sc.addPyFile("/eos/home-d/dalfonso/SWAN_projects/Hrare/JULY_exp/utilsAna.py")
df = (dfINI. .Define("myInt","ROOT.CountCharacters('Hello, let me try again today.')"))
with the simple function like this
[dalfonso@jupyter-dalfonso ~]$ cat /eos/home-d/dalfonso/SWAN_projects/Hrare/JULY_exp/myLibrary.h
#include <iostream>
#include <typeinfo>
/// A trivial function
int CountCharacters(const std::string s)
{
return s.size();
}
I can print the
ROOT.CountCharacters(“Hello, let me try again today.”)
on a notebook shell, but when I have the define of RDF
return rdf._OriginalDefine(col_name, callable_or_str)
cppyy.gbl.std.runtime_error: Template method resolution failed:
ROOT::RDF::RInterface<ROOT::Detail::RDF::RJittedFilter,void> ROOT::RDF::RInterface<ROOT::Detail::RDF::RJittedFilter,void>::Define(basic_string_view<char,char_traits<char> > name, basic_string_view<char,char_traits<char> > expression) =>
runtime_error:
RDataFrame: An error occurred during just-in-time compilation. The lines above might indicate the cause of the crash
All RDF objects that have not run an event loop yet should be considered in an invalid state.
ROOT::RDF::RInterface<ROOT::Detail::RDF::RJittedFilter,void> ROOT::RDF::RInterface<ROOT::Detail::RDF::RJittedFilter,void>::Define(basic_string_view<char,char_traits<char> > name, basic_string_view<char,char_traits<char> > expression) =>
runtime_error:
RDataFrame: An error occurred during just-in-time compilation. The lines above might indicate the cause of the crash
All RDF objects that have not run an event loop yet should be considered in an invalid state.