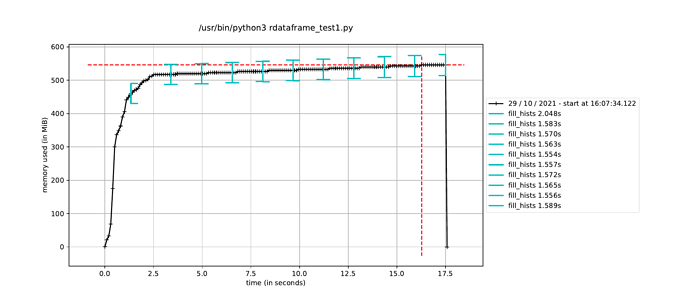
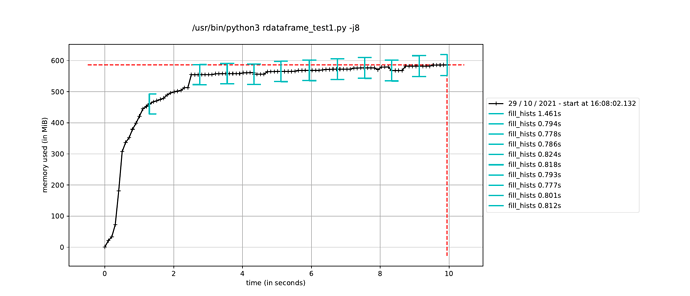
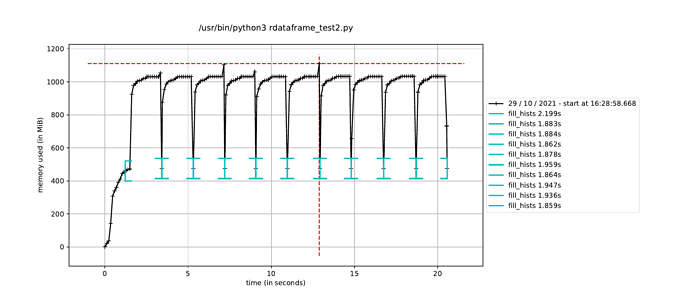
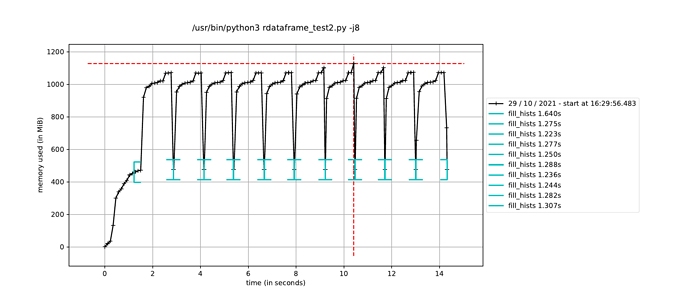
Hi there,
I’m using RDataFrames in PyROOT for my analysis to read in ntuples and fill a number of histograms. In most cases I use a pre-compiled library to apply my filters and new-column definitions, however in some case it was more convenient to apply these actions as string expressions, resulting in just-in-time compilations. Because of the large numbers of samples I have, this JITing resulted in the memory-hog issue described elsewhere, e.g. JIT Compilation Memory Hogs in RDataFrame.
To fix this memory issue, I run the RDataFrame event loop in a new process so that all the memory consumed by the JIT compilation is released once the event loop finishes. I define the histograms in the parent process and trigger the event in the child process. This is a condensed version of what I am doing:
import ROOT
import multiprocessing
ROOT.ROOT.EnableImplicitMT()
def fill_hists(df, hist_args):
df_local = df.Filter("...", "...").Define("...", "...")
rhists = []
for args in hist_args:
rhists.append(df_local.Histo1D(*args))
with multiprocessing.Manager() as manager:
results = manager.dict()
proc = multiprocessing.Process(
target=fill_hists_worker,
args=(results, rhists)
)
proc.start()
proc.join()
def fill_hists_worker(results, rhists):
for rhist in rhists:
hist = rdf_hist.GetValue()
results[hist.GetName()] = hist
However, when I trigger the event loop in a new process in this way, the execution time to fill the histograms increases by about a factor of how many CPU cores I have, leading me to believe it is running the event loop in a single thread dispite having called ROOT.ROOT.EnableImplicitMT(). The alternative, as mentioned elsewhere, is to using a multiprocessing.Pool() instead of a single process, but I would have thought this would run a separate event loop in each process in the pool, which is not ideal since only a single event loop is necessary.
Is it possible to use ROOT’s implicit MT in a Python multiprocessing.Process()? And if not, what would the solution be to run a single event loop in parallel while still avoiding the JIT memory-hogging issue?
NOTE: I am using ROOT v6.20/04, since this is the latest version available on my local cluster.
Thanks,
Joey
ROOT Version: 6.20/04
Platform: x86_64 (Red Hat 4.8.5-44)
Compiler: gcc 9.3.0