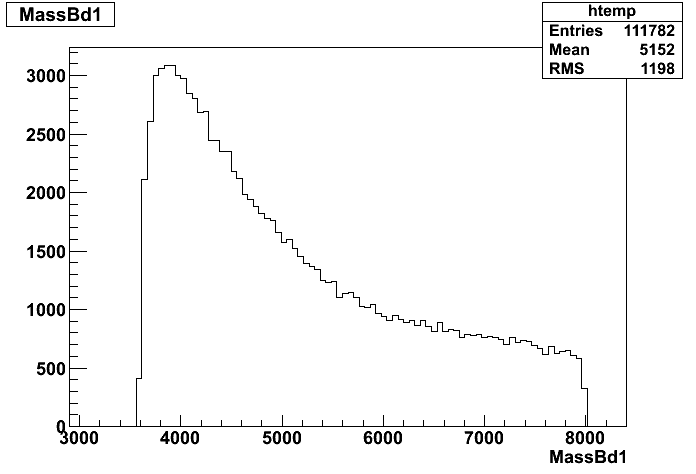
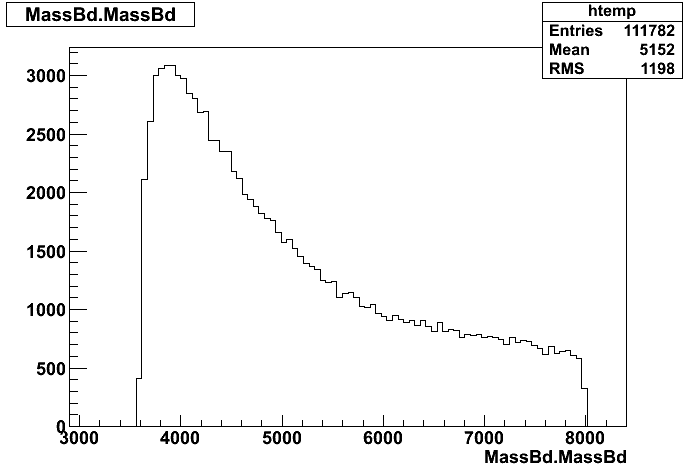
Dear Rooters,
I am interesting to fit some of my ntuple branches using rooFit, but all my branches are type vectors and I try to transfer them from vectors to double to use the roofit functions, to do that i am using a macro but it doesn’t works well:
#include ;
void tree5() {
//Get old file, old tree and set top branch address
TFile *f = new TFile("/tmp/gouighri/MergentupleCDF_E2.root",“READ”);
TTree inputtree = (TTree)f->Get(“BmesonntupleCDF”);
vector *pvec1(0);
vector *pvec2(0);
vector *pvec3(0);
vector *pvec4(0);
inputtree->SetBranchAddress(“MassBd”,&pvec1);
inputtree->SetBranchAddress(“TauBd”,&pvec2);
inputtree->SetBranchAddress(“LxyBd”,&pvec3);
inputtree->SetBranchAddress(“LxyJFit”,&pvec4);
TFile *outputfile = new TFile("/tmp/gouighri/test1.root",“recreate”);
TTree *outputtree = new TTree(“tree”,“tree with doubles”);
double MassBd1;
double TauBd1;
double LxyBd1;
double LxyJFit1;
outputtree->Branch(“MassBd1”,&MassBd1);
outputtree->Branch(“TauBd1”,&TauBd1);
outputtree->Branch(“LxyBd1”,&LxyBd1);
outputtree->Branch(“LxyJFit1”,&LxyJFit1);
for(Long64_t entry = 0; entry < inputtree->GetEntries(); ++entry) {
inputtree->GetEntry(entry);
std::vector::iterator iter1 = pvec1->begin();
std::vector::iterator iter2 = pvec2->begin();
std::vector::iterator iter3 = pvec3->begin();
std::vector::iterator iter4 = pvec4->begin();
assert( pvec1->size() == pvec2->size() && pvec2->size() == pvec3->size() && pvec2->size() == pvec4->size() );
for( ; iter1 != pvec1->end(); ++iter1, ++iter2, ++iter3, ++iter4) {
MassBd1 = (*iter1);
TauBd1 = (*iter2);
LxyBd1 = (*iter3);
LxyJFit1 = (*iter4);
outputtree->Fill();
}
}
outputfile->Write();
outputfile->Close();
}
Please do you have an idea what i am doing wrong? How can I manage it?
Thank you very much for your help
Cheers
Mohamed