For the question1; is it mandatory to recompile root to have the chunk option?
( I am not allowed to recompile root…)
No, the chunk option should already be available, my patch is only a performance optimization.
Thanks enrico for your answer
I adapted my pgm to build an exe
But when I run it , it crashes
vinchon-thi@cadux-login01:/CADUX-DESTO/SDOS/vg2022/Van_Gogh_22g/DAQ/Ron35/UNFILTERED$ ./exe*/CSV_to_root_viaRDF
avt appel CSV_to_root_viaRDF()
apres MakeCsvDataFrame
*** Error in `./execLinux64/CSV_to_root_viaRDF’: free(): invalid pointer: 0x0000000004789e98 ***
I do not understand from where come this invalid pointer…
See below the pgm
*’’#include “Riostream.h”
#include “TString.h”
#include “TFile.h”
#include “TTree.h”
#include “TCanvas.h”
#include “/soft/root/6.22.08/include/ROOT/RDataFrame.hxx”
#include “/soft/root/6.22.08/include/ROOT/RCsvDS.hxx”
#include “TSystem.h”****int CSV_to_root_viaRDF()
{
ROOT::EnableImplicitMT();
// Let’s first create a RDF that will read from the CSV file.
auto df = ROOT::RDF::MakeCsvDataFrame(“Data_CH6@DT5730S_10548_Ron35.csv”);***
std::cout << “apres MakeCsvDataFrame”<<std::endl;
df.Snapshot(“myTree”, “Data_CH6@DT5730S_10548_Ron35.root”);***
std::cout << “apres Snapshot”<<std::endl;
return 0;
}
int main( int argc, char* argv[] )
{
std::cout<< “avt appel CSV_to_root_viaRDF()”<<std::endl;
CSV_to_root_viaRDF();***
std::cout<< “apres appel CSV_to_root_viaRDF()”<<std::endl;
}
ROOT::RDF::MakeCsvDataFrame(“Data_CH6@DT5730S_10548_Ron35.csv”); should be ROOT::RDF::MakeCsvDataFrame("Data_CH6@DT5730S_10548_Ron35.csv", true, ';', 1000000); as discussed above, other than that I don’t see any problems.
I can run execute this program on a CSV file like yours and it works:
#include "ROOT/RCsvDS.hxx"
#include "ROOT/RDataFrame.hxx"
#include "Riostream.h"
#include "TCanvas.h"
#include "TFile.h"
#include "TString.h"
#include "TSystem.h"
#include "TTree.h"
int CSV_to_root_viaRDF() {
ROOT::EnableImplicitMT();
// Let’s first create a RDF that will read from the CSV file.
auto df = ROOT::RDF::MakeCsvDataFrame("./large_body.csv", true, ';', 1000000);
std::cout << "apres MakeCsvDataFrame" << std::endl;
df.Snapshot("myTree", "out.root");
std::cout << "apres Snapshot" << std::endl;
return 0;
}
int main(int argc, char *argv[]) {
std::cout << "avt appel CSV_to_root_viaRDF()" << std::endl;
CSV_to_root_viaRDF();
std::cout << "apres appel CSV_to_root_viaRDF()" << std::endl;
}
Maybe the worker is just running out of memory because of the lack of chunking?
Enrico
If I add the chunk argument as you proposed I get a warning at compilation ( and the same crash after…)
- compilation of CSV_to_root_viaRDF.cc
main/CSV_to_root_viaRDF.cc: In function ‘int CSV_to_root_viaRDF()’:
main/CSV_to_root_viaRDF.cc:16:86: warning: overflow in implicit constant conversion [-Woverflow]
auto df = ROOT::RDF::MakeCsvDataFrame(“Data_CH6@DT5730S_10548_Ron35.csv”,’;’,10000);
Hi Enrico
For information, the latest version we have on the server is ROOT 6.22/08
Can you repeat your test with this version
Regards
Thibaut
The compiler warning is correct, the code above is wrong. It should be ROOT::RDF::MakeCsvDataFrame(“Data_CH6@DT5730S_10548_Ron35.csv”, true, ’;’, 10000);, see ROOT: ROOT::RDF Namespace Reference .
Cheers,
Enrico
I just tested reading and writing out a CSV of 11GB with your same schema with ROOT v6.22 installed via conda, on my laptop. It took around 20 minutes:
$ conda activate cern-root-622
$ root-config --version
6.22/08
$ cat program.cpp
#include "ROOT/RCsvDS.hxx"
#include "ROOT/RDataFrame.hxx"
#include "Riostream.h"
#include "TCanvas.h"
#include "TFile.h"
#include "TString.h"
#include "TSystem.h"
#include "TTree.h"
int CSV_to_root_viaRDF() {
ROOT::EnableImplicitMT();
// Let’s first create a RDF that will read from the CSV file.
auto df = ROOT::RDF::MakeCsvDataFrame("./large_body.csv", true, ';', 1000000);
std::cout << "apres MakeCsvDataFrame" << std::endl;
df.Snapshot("myTree", "out.root");
std::cout << "apres Snapshot" << std::endl;
return 0;
}
int main(int argc, char *argv[]) {
std::cout << "avt appel CSV_to_root_viaRDF()" << std::endl;
CSV_to_root_viaRDF();
std::cout << "apres appel CSV_to_root_viaRDF()" << std::endl;
}
$ rootcompile program.cpp -O3 && /usr/bin/time ./program
g++ -g -Wall -Wextra -Wpedantic -o "program" "program.cpp" $(root-config --cflags --libs) -O3
program.cpp: In function 'int main(int, char**)':
program.cpp:20:14: warning: unused parameter 'argc' [-Wunused-parameter]
20 | int main(int argc, char *argv[]) {
| ~~~~^~~~
program.cpp:20:26: warning: unused parameter 'argv' [-Wunused-parameter]
20 | int main(int argc, char *argv[]) {
| ~~~~~~^~~~~~
avt appel CSV_to_root_viaRDF()
apres MakeCsvDataFrame
apres Snapshot
apres appel CSV_to_root_viaRDF()
1995.05user 8.70system 20:38.01elapsed 161%CPU (0avgtext+0avgdata 943652maxresident)k
1328inputs+1362400outputs (25major+379668minor)pagefaults 0swaps
Despite the EnableImplicitMT the program mostly runs single-threaded because almost all the time is spent in parsing the CSV, which is a sequential operation, rather than data processing which is what is parallelized.
With the performance optimization in my patch above runtimes are down to around 5 minutes for that same input file.
Cheers,
Enrico
P.S. these times should scale linearly with the CSV size, so on my laptop I would expect a bit more than 3x those runtimes for your real input of almost 40 GB.
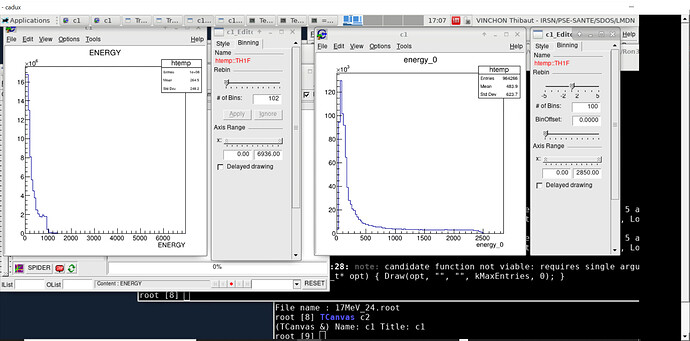
Dear Enrico
In both methods , the histgram buid with the treeviewer have a very small number of bins ( 102 for a energy for example and 51 for a bidim energy:energy_short)
On the left, an histogram build from the CSV_to_root_viaRDF() program:
number of bin cannot be increased
On the right a root file build by another way
number of bin can be increase
Is it related to the big number of entries of the tree?
Regards
Thibaut
You can control the number of bins and the min-max range, using the TreeViewer and then htemp(200,0,3000) as draw option.
If you want to always change the default value of 100, see TTree::Draw -> how to change the default number of bins?
thanks for this it works
Regards
Thibaut
Dear Enrico
it still doesnt work
Note that the violation is detected at the snapshot and not before…
note that the csv size is 35G0 ( and could be 59G0 for other files)
11G0 is the size of the root file obtein with the ReadFile method…
Hi @Thibaut_VINCHON ,
I cannot reproduce the problem, as I mentioned I could process a large CSV without issues with RDF+Snapshot on my laptop. Can you share a complete recipe (including data) to reproduce the problem e.g. on lxplus or in a Docker container? If not, can you attach gdb and print the stacktrace at the point of crash? For this to be useful you will need a ROOT version compiled with debug symbols, available e.g. from LXPLUS.
About the fact that the crash happens at the line where you call Snapshot: that’s the line at which the full event loop is run, so the crash happens somewhere during the data processing but without a full stacktrace it’s hard to tell where or why.
For the histogram binning, note that you can tell RDataFrame what binning you want passing a histogram or a histogram model with the desired binning to Histo1D, see e.g. ROOT: ROOT::RDF::RInterface< Proxied, DataSource > Class Template Reference .
Cheers,
Enrico
for “fun”, I tried to implement this with Go-HEP/groot.
here is the Linux binary:
-
https://cern.ch/binet/go-hep/csv2root-irsn.exe
(just download it to a Linux machine, and make it executable:chmod +x ./csv2root-irsn.exe)
it should work on any Linux machine:
$> ./csv2root -o out.root -t irsn ./data.csv
csv2root: read CSV header: ["BOARD" "CANAL" "TIMETAG" "ENERGY" "ENERGY_SHORT" "FLAG"]
csv2root: handled 8 events
$> root-dump ./out.root
>>> file[./out.root]
key[000]: irsn;1 "" (TTree)
[000][Board]: 0
[000][Channel]: 2
[000][Timetag]: 2
[000][Energy]: 395
[000][EneShort]: 255
[000][Flag]: 16384
[001][Board]: 0
[001][Channel]: 2
[001][Timetag]: 2
[001][Energy]: 512
[...]
I’d be interested to know whether this fits your bill ![]()
This topic was automatically closed 14 days after the last reply. New replies are no longer allowed.