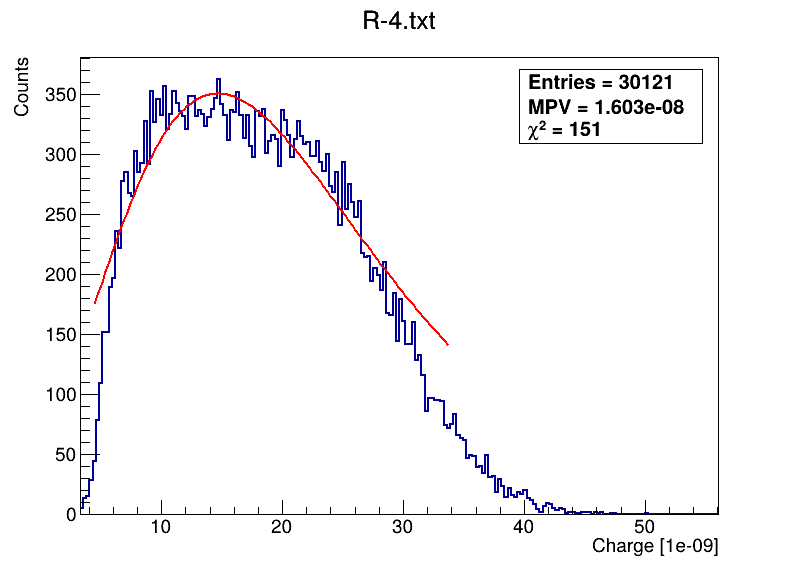
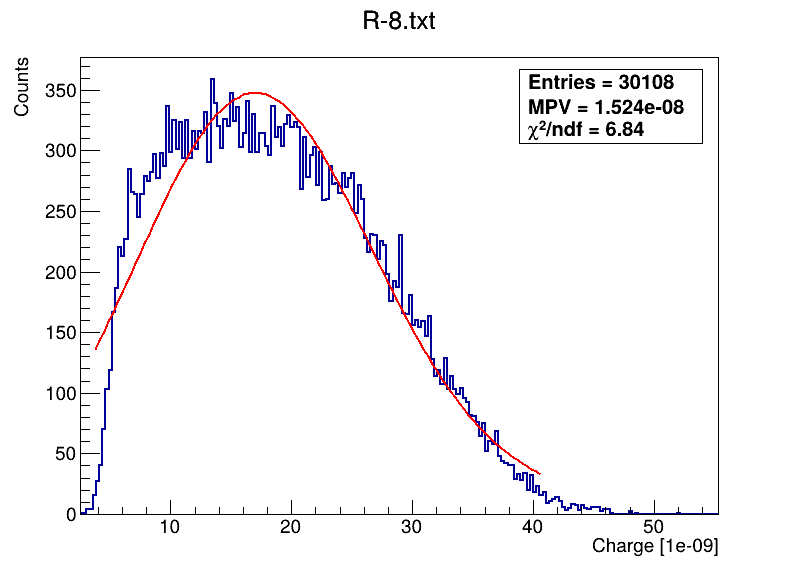
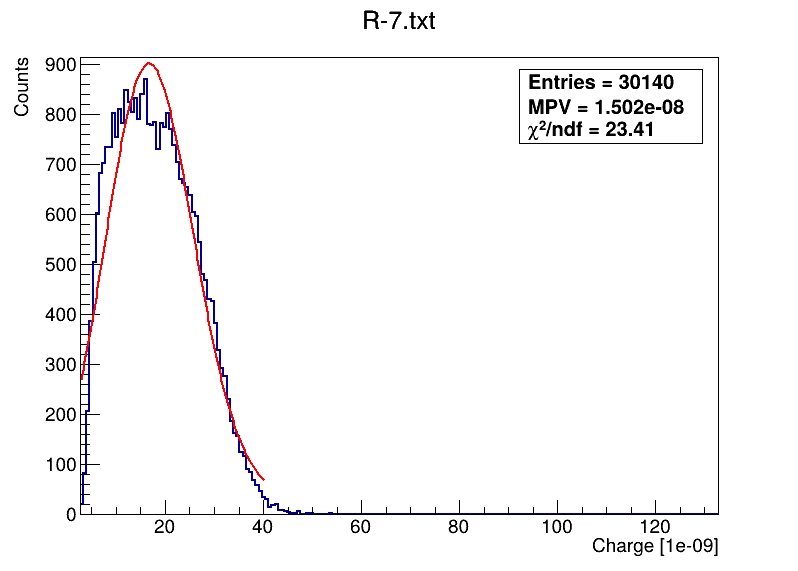
This is the code what I am using:
#include "TSystemDirectory.h"
#include "TSystemFile.h"
#include "TList.h"
#include "TCollection.h"
#include "TCanvas.h"
#include "TH1D.h"
#include "TF1.h"
#include "TString.h"
#include "TROOT.h"
#include "TStyle.h"
#include "TMath.h"
#include "TGraphErrors.h"
#include "TLegend.h"
#include "TPaveText.h"
#include <fstream>
#include <iostream>
#include <vector>
#include <string>
#include <limits>
#include <map>
#include <sstream>
#include <cmath>
// global path for CSV with fit results
static TString gResultsPath;
// --------------------------------------------------------
// Langau: Landau ⊗ Gaussian convolution
// par[0] = Landau width
// par[1] = MPV
// par[2] = Area (overall norm)
// par[3] = Gaussian sigma
// --------------------------------------------------------
Double_t langaufun(Double_t *x, Double_t *par)
{
// numeric convolution
const Double_t invsq2pi = 0.3989422804014327; // 1 / sqrt(2*pi)
const Double_t np = 100.0; // number of steps
const Double_t sc = 5.0; // convolution range in sigma
Double_t xx = x[0];
Double_t mpc = par[1];
Double_t width = par[0];
Double_t sigma = par[3];
Double_t area = par[2];
Double_t xlow = xx - sc*sigma;
Double_t xupp = xx + sc*sigma;
Double_t step = (xupp - xlow)/np;
Double_t sum = 0.0;
Double_t xval = xlow + 0.5*step;
for (int i = 0; i < np; ++i) {
Double_t land = TMath::Landau(xval, mpc, width, true); // normalized
Double_t arg = (xx - xval)/sigma;
Double_t gaus = TMath::Exp(-0.5*arg*arg);
sum += land * gaus;
xval += step;
}
return area * step * invsq2pi / sigma * sum;
}
// --------------------------------------------------------
// Read amplitudes and convert to "nano units":
// An = A / 1e-9 (so x-axis is 0–60 instead of 0–6e-8)
// --------------------------------------------------------
bool ReadAmplitudesNano(const std::string &filepath, std::vector<double> &sNano)
{
std::ifstream in(filepath.c_str());
if (!in.is_open()) {
std::cerr << "Cannot open file: " << filepath << std::endl;
return false;
}
std::string line;
bool dataSection = false;
while (std::getline(in, line)) {
if (line.find("Time Ampl") != std::string::npos) {
dataSection = true;
break;
}
}
if (!dataSection) {
std::cerr << "Did not find 'Time Ampl' in file: " << filepath << std::endl;
return false;
}
long long index;
double ampl;
while (in >> index >> ampl) {
ampsNano.push_back(ampl / 1e-9); // "nano" units
}
if (ampsNano.empty()) {
std::cerr << "No amplitude data in file: " << filepath << std::endl;
return false;
}
return true;
}
void FitPeakLangauAndSave(TH1D *h, const TString &fnameTxt, const TString &pngPath)
{
if (!h) return;
gStyle->SetOptStat(0);
gStyle->SetOptFit(0);
double xmin = h->GetXaxis()->GetXmin();
double xmax = h->GetXaxis()->GetXmax();
int nbins = h->GetNbinsX();
int maxBin = h->GetMaximumBin();
double mpv0 = h->GetBinCenter(maxBin);
double maxCnt = h->GetMaximum();
// ----------------------------------------------------
// 1) Define a "core" region: bins above some fraction
// of the maximum (peak area only).
// Tune CORE_FRAC if you want narrower/wider fits.
// ----------------------------------------------------
const double CORE_FRAC = 0.40; // try 0.4–0.5
double thrCore = CORE_FRAC * maxCnt;
int leftCoreBin = maxBin;
int rightCoreBin = maxBin;
while (leftCoreBin > 1 && h->GetBinContent(leftCoreBin) > thrCore)
--leftCoreBin;
while (rightCoreBin < nbins && h->GetBinContent(rightCoreBin) > thrCore)
++rightCoreBin;
double fitMin = h->GetBinLowEdge(leftCoreBin);
double fitMax = h->GetBinLowEdge(rightCoreBin + 1);
if (fitMin < xmin) fitMin = xmin;
if (fitMax > xmax) fitMax = xmax;
// ----------------------------------------------------
// 2) Estimate "core RMS" only from the peak region,
// to get better starting values.
// ----------------------------------------------------
double sum = 0.0;
double sumx = 0.0;
double sumx2 = 0.0;
for (int bin = leftCoreBin; bin <= rightCoreBin; ++bin) {
double c = h->GetBinContent(bin);
double xc = h->GetBinCenter(bin);
sum += c;
sumx += c * xc;
sumx2 += c * xc * xc;
}
double meanCore = (sum > 0.0) ? sumx / sum : mpv0;
double varCore = (sum > 0.0) ? (sumx2 / sum - meanCore*meanCore) : 0.0;
double rmsCore = (varCore > 0.0) ? std::sqrt(varCore) : (xmax - xmin)/20.0;
double width0 = 0.7 * rmsCore;
double sigma0 = 0.7 * rmsCore;
double area0 = sum; // ~ area under the core
// ----------------------------------------------------
// 3) Langau fit, restricted to the core region
// ----------------------------------------------------
TF1 *f = new TF1("f_langG", langaufun, fitMin, fitMax, 4);
f->SetParNames("Width","MPV","Area","Sigma");
f->SetParameters(width0, mpv0, area0, sigma0);
f->SetParLimits(0, 0.1 * rmsCore, 3.0 * rmsCore); // width
f->SetParLimits(1, mpv0 - 1.0*rmsCore, mpv0 + 1.0*rmsCore); // MPV
f->SetParLimits(2, 0.01 * area0, 1e5 * area0); // area
f->SetParLimits(3, 0.1 * rmsCore, 3.0 * rmsCore); // sigma
TFitResultPtr r = h->Fit(f, "RQS"); // R: use range, Q: quiet, S: store
// OPTIONAL: one retry if it failed badly
if (r->Status() != 0) {
f->SetParameters(width0, mpv0, area0, sigma0);
h->Fit(f, "RQB"); // fallback fit
}
// ----------------------------------------------------
// 4) Draw and save – we keep the SAME (peak-only) range
// so the red curve stops before the long tail.
// ----------------------------------------------------
TCanvas *c = new TCanvas(TString(h->GetName()) + "_c",
TString(h->GetName()) + "_c",
800, 600);
h->GetXaxis()->SetTitle("Charge [1e-09]");
h->GetYaxis()->SetTitle("Counts");
h->SetLineWidth(2);
h->Draw("HIST");
f->SetRange(fitMin, fitMax); // NO extension into the tail
f->SetLineColor(kRed);
f->SetLineWidth(2);
f->Draw("SAME");
double entries = h->GetEntries();
double mpv_scaled = f->GetParameter(1);
double mpv_phys = mpv_scaled * 1e-9;
double chi2 = f->GetChisquare();
TPaveText *pt = new TPaveText(0.65, 0.75, 0.88, 0.88, "NDC");
pt->SetFillColor(0);
pt->SetBorderSize(1);
pt->SetTextAlign(12);
pt->AddText(Form("Entries = %.0f", entries));
pt->AddText(Form("MPV = %.3e", mpv_phys));
pt->AddText(Form("#chi^{2} = %.3g", chi2));
pt->Draw();
TString pngName = pngPath + "/" + TString(h->GetName()) + "_peak.png";
c->SaveAs(pngName);
// Save results to CSV
std::ofstream ofs(fnameTxt.Data(), std::ios::app);
ofs << h->GetName() << ","
<< mpv_phys << ","
<< entries << ","
<< chi2 << "\n";
ofs.close();
delete c;
}
// --------------------------------------------------------
// Make normalized MPV vs distance plot with exponential
// fits for left (L-*) and right (R-*).
// --------------------------------------------------------
void PlotNormalized(const char *dirpath = ".")
{
TString csvPath = TString::Format("%s/fit_results.csv", dirpath);
std::ifstream in(csvPath.Data());
if (!in.is_open()) {
std::cerr << "Cannot open results file: " << csvPath << std::endl;
return;
}
std::string header;
std::getline(in, header); // skip header
struct Row {
std::string fname;
double mpv = 0.0; // physical units
double entries = 0.0;
double chi2 = 0.0;
};
std::map<int, Row> left, right;
double mpvLref = -1.0; // MPV of L-1
double mpvRref = -1.0; // MPV of R-9
std::string line;
while (std::getline(in, line)) {
if (line.empty()) continue;
std::stringstream ss(line);
std::string fname, s_mpv, s_entries, s_chi2;
std::getline(ss, fname, ',');
std::getline(ss, s_mpv, ',');
std::getline(ss, s_entries, ',');
std::getline(ss, s_chi2, ',');
Row r;
r.fname = fname;
r.mpv = std::atof(s_mpv.c_str());
r.entries = std::atof(s_entries.c_str());
r.chi2 = std::atof(s_chi2.c_str());
if (fname.size() < 3) continue;
char side = fname[0];
std::size_t dashPos = fname.find('-');
if (dashPos == std::string::npos) continue;
std::size_t dotPos = fname.find('.', dashPos + 1);
if (dotPos == std::string::npos) dotPos = fname.size();
int idx = std::atoi(fname.substr(dashPos + 1,
dotPos - dashPos - 1).c_str());
if (idx <= 0) continue;
if (side == 'L') {
left[idx] = r;
if (idx == 1) mpvLref = r.mpv;
} else if (side == 'R') {
right[idx] = r;
if (idx == 9) mpvRref = r.mpv;
}
}
in.close();
if (left.empty() && right.empty()) {
std::cerr << "No L-* or R-* entries in results file." << std::endl;
return;
}
int nL = left.size();
int nR = right.size();
std::vector<double> xL(nL), yL(nL), exL(nL, 0.3), eyL(nL);
std::vector<double> xR(nR), yR(nR), exR(nR, 0.3), eyR(nR);
int i = 0;
for (auto &kv : left) {
int idx = kv.first;
const Row &r = kv.second;
xL[i] = static_cast<double>(idx); // distance in cm
yL[i] = (mpvLref > 0.0) ? (r.mpv / mpvLref) : 0; // normalized
eyL[i] = (r.entries > 0.0) ? 1.0 / std::sqrt(r.entries) : 0.0;
++i;
}
i = 0;
for (auto &kv : right) {
int idx = kv.first;
const Row &r = kv.second;
xR[i] = static_cast<double>(idx);
yR[i] = (mpvRref > 0.0) ? (r.mpv / mpvRref) : 0;
eyR[i] = (r.entries > 0.0) ? 1.0 / std::sqrt(r.entries) : 0.0;
++i;
}
double maxX = 0.0;
if (!xL.empty())
maxX = std::max(maxX, *std::max_element(xL.begin(), xL.end()));
if (!xR.empty())
maxX = std::max(maxX, *std::max_element(xR.begin(), xR.end()));
if (maxX <= 0) maxX = 10.0;
// no default fit box here
gStyle->SetOptFit(0);
TCanvas *c = new TCanvas("c_norm", "Normalized MPV vs distance", 800, 600);
TGraphErrors *gL = nullptr;
TGraphErrors *gR = nullptr;
if (nL > 0) {
gL = new TGraphErrors(nL, xL.data(), yL.data(), exL.data(), eyL.data());
gL->SetName("gLeft");
gL->SetTitle("");
gL->SetMarkerStyle(20);
gL->SetMarkerColor(kBlue+1);
gL->SetLineColor(kBlue+1);
gL->Draw("AP");
gL->GetXaxis()->SetTitle("Distance [cm]");
gL->GetYaxis()->SetTitle("MPV / reference MPV");
gL->GetYaxis()->SetRangeUser(0.75, 1.05);
// exponential fit: y = A * exp(-x / lambda)
TF1 *fL = new TF1("fL", "[0]*TMath::Exp(-x/[1])", 0.0, maxX+1.0);
fL->SetParameters(1.0, 10.0);
fL->SetLineColor(kBlue+2);
fL->SetLineWidth(2);
if (nL >= 2) gL->Fit(fL, "RQ0"); // quiet, no auto-draw box
fL->Draw("SAME");
}
if (nR > 0) {
gR = new TGraphErrors(nR, xR.data(), yR.data(), exR.data(), eyR.data());
gR->SetName("gRight");
gR->SetMarkerStyle(21);
gR->SetMarkerColor(kRed+1);
gR->SetLineColor(kRed+1);
if (gL) gR->Draw("P SAME");
else {
gR->Draw("AP");
gR->GetXaxis()->SetTitle("Distance [cm]");
gR->GetYaxis()->SetTitle("MPV / reference MPV");
gR->GetYaxis()->SetRangeUser(0.75, 1.05);
}
TF1 *fR = new TF1("fR", "[0]*TMath::Exp(-x/[1])", 0.0, maxX+1.0);
fR->SetParameters(1.0, 10.0);
fR->SetLineColor(kRed+2);
fR->SetLineWidth(2);
if (nR >= 2) gR->Fit(fR, "RQ0");
fR->Draw("SAME");
}
TLegend *leg = new TLegend(0.60, 0.70, 0.88, 0.88);
if (gL) leg->AddEntry(gL, "Left (L-*)", "lp");
if (gR) leg->AddEntry(gR, "Right (R-*)", "lp");
leg->Draw();
c->SaveAs(TString::Format("%s/mpv_vs_distance.png", dirpath));
delete c;
delete gL;
delete gR;
delete leg;
}
// --------------------------------------------------------
// Main entry: loop over *.txt, make histograms, fit, save,
// write fit_results.csv, then call PlotNormalized().
// --------------------------------------------------------
void plot(const char *dirpath)
{
// prepare CSV with header
gResultsPath = TString::Format("%s/fit_results.csv", dirpath);
{
std::ofstream out(gResultsPath.Data());
if (!out.is_open()) {
std::cerr << "Cannot open results file for writing: "
<< gResultsPath << std::endl;
} else {
out << "filename,mpv,entries,chi2\n";
}
}
TSystemDirectory dir("dataDir", dirpath);
TList *files = dir.GetListOfFiles();
if (!files) {
std::cerr << "No files found in directory: " << dirpath << std::endl;
return;
}
TIter next(files);
TSystemFile *file;
while ((file = (TSystemFile *)next())) {
TString fname = file->GetName();
if (file->IsDirectory()) continue;
if (fname.BeginsWith(".")) continue;
if (!fname.EndsWith(".txt")) continue;
if (!(fname.BeginsWith("L-") || fname.BeginsWith("R-"))) continue;
TString fullpath = TString::Format("%s/%s", dirpath, fname.Data());
std::cout << "Processing: " << fullpath << std::endl;
std::vector<double> ampsNano;
if (!ReadAmplitudesNano(fullpath.Data(), ampsNano)) {
std::cerr << "Skipping file: " << fullpath << std::endl;
continue;
}
auto mm = std::minmax_element(ampsNano.begin(), ampsNano.end());
double xmin = *mm.first;
double xmax = *mm.second;
if (xmin == xmax) { xmin -= 0.5; xmax += 0.5; }
TString histName = fname;
histName.ReplaceAll(".txt", "_h");
TH1D *h = new TH1D(histName, fname, 200, xmin, xmax);
for (double a : ampsNano) h->Fill(a);
TString outpng = fname;
outpng.ReplaceAll(".txt", "_langaus.png");
outpng = TString::Format("%s/%s", dirpath, outpng.Data());
FitPeakLangauAndSave(h, fname, outpng);
delete h;
}
// normalized exponential plot
PlotNormalized(dirpath);
}
// overload with no args so ROOT can auto-run when you do: root -l plot.C
void plot()
{
plot(".");
}