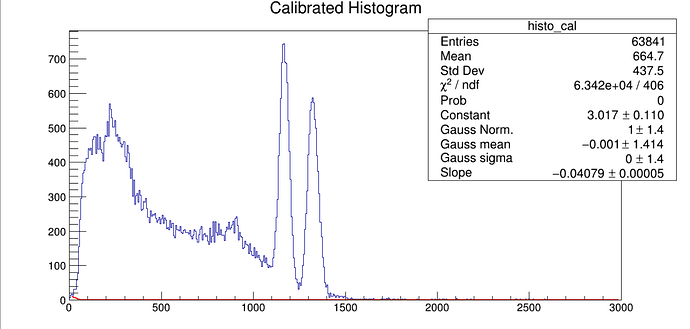
Hello,
I am trying to perform a gaussian fit of a photopeak for several elements. I also want to include an exponential background noise but I have trouble in defining the final fit function. The Gaussian fit actually works fine but when I combine the two something weird shows up (see attached file).
#include "TCanvas.h"
#include "TFile.h"
#include "TH1.h"
#include "TF1.h"
#include "TAxis.h"
#include "TStyle.h"
#include <string>
#include <iostream>
void analisidati_gamma1(
std::string filename="Co60hq0.root"
,std::string histoname="hq0"
,std::string bkgfunc="expo"
,int xminGFit1=1120
,int xmaxGFit1=1240
//,int xminGFit2=1250
//,int xmaxGFit2=1400
){
gStyle->SetOptFit(1111);
TCanvas* can = new TCanvas("can","my canvas",800,800);
// retrieve the histogram from the input file
TFile* file = TFile::Open(filename.c_str());
if(!file){
std::cout << "File " << filename << " not found... check the input name!" << std::endl;
return;
}
TH1D* histo = (TH1D*) file->Get(histoname.c_str());
if(!histo){
std::cout << "histo " << histoname << " not found... check the input name!" << std::endl;
return;
}
// Get the TTree from the file
TTree *nt = (TTree*) file->Get("nt");
// Create a new histogram with 500 bins in the range [0, 10000]
TH1D *histonew = new TH1D("histonew", "hq0new", 500, 0, 10000);
// Draw the variable "qlong" from the TTree "nt" into the histogram "histonew"
nt->Draw("qlong>>histonew", "ch==0");
TH1D* histo_copy = (TH1D*)histonew->Clone();
histo_copy->SetName("histo_copy");
// Calibration
TH1D *histo_cal = new TH1D("histo_cal", "Calibrated Histogram", 500, 0, 3000);
nt->Draw("-32.4+qlong*0.1942>>histo_cal", "ch==0");
// Gaussian fit
const double normGaus=100; // Gauss normalization
const double meanGaus=1700; // Gauss mean
const double sigmaGaus=500;
// fit function
std::string fitfunc = "gaus(0)";
TF1* gaussFunc = new TF1("gaussFunc",fitfunc.c_str(),0,1000);
gaussFunc->SetParName(0,"Gauss Norm.");
gaussFunc->SetParName(1,"Gauss mean");
gaussFunc->SetParName(2,"Gauss sigma");
gaussFunc->SetParameter(100,normGaus);
gaussFunc->SetParameter(1170,meanGaus);
gaussFunc->SetParameter(500,sigmaGaus);
//histo_cal->Fit(myfit,"","",xminGFit1,xmaxGFit1);
//TFile* file_output = new TFile("file_output.root","recreate");
//can->Write();
//histo_cal->Write();
// Define background exponential function
TF1 *bkgFunc = new TF1("bkgFunc", "expo", 0, 3000);
// Combine Gaussian and background functions
TF1 *fitFunc = new TF1("fitFunc", "gaussFunc(2) + bkgFunc", xminGFit1, xmaxGFit1);
// Set initial parameters for background exponential function
bkgFunc->SetParameters(0, 1.0); // Exponential amplitude
bkgFunc->SetParameters(1, -0.001); // Exponential decay constant
// Set initial parameters for combined function
fitFunc->SetParameters(normGaus, 1.0, -0.001);
// Fit combined function to histogram
histo_cal->Fit(fitFunc, "", "", 0, 3000);
return;
}