I would like to perform a sort of ensemble test by fitting a gaussian PDF to gaussian generated data. I have coded up something but for some reason my pull distributions are all off. The following with code is what I have done.
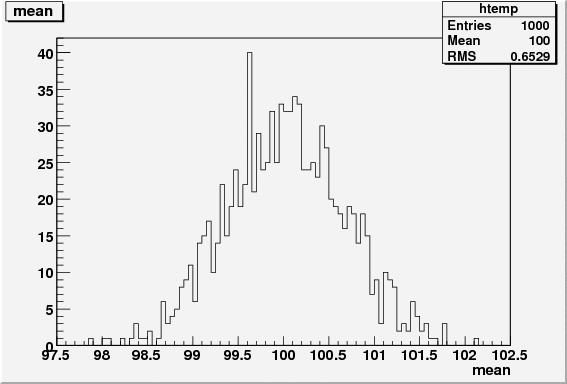
1: Generate a gaussian distribution with a mean of 100 and a width of 20 (I should specifiy that nentries = 1000 here)
TH1* genMC(TRandom3 *rng, TH1 *input) {
if(input == 0x0) return 0x0;
// Clone the histogram
TH1 *output = (TH1*) input->Clone();
long nentries = output->GetEntries();
// Empty it out
output->Reset();
for(long i = 0; i < nentries; i++) {
output->Fill(rng->Gaus(100, 20));
}
return(output);
}2: I then pass this into a function that does the fitting using RooFit
Result doFit(TH1 *input_sideband, TH1 *input_signal, bool saveme) {
Result res;
double signal_region_count = input_signal->Integral(input_signal->FindBin(0), input_signal->FindBin(300));
double sideband_region_count = input_sideband->Integral(input_sideband->FindBin(0), input_sideband->FindBin(300));
RooRealVar gauss_mean("mean", "mean", 70, 50, 150);
RooRealVar gauss_width("width", "width", 15, 0, 30);
RooGaussian thresh("gauss", "Signal PDF", x, gauss_mean, gauss_width);
RooDataHist data_sideband("data", "Yhea", x, input_sideband);
x.setRange("fit", 0, 300);
RooPlot *frame = 0x0;
frame = x.frame();
data_sideband.plotOn(frame, MarkerColor(1), LineColor(1));
data_sideband.statOn(frame);
// Generate the extended likelyhood stuff
RooRealVar nbkg("nbkg", "Number of Background Events", 100, 0, 10000000);
RooAddPdf bkgModel("BkgModel", "Background Model", RooArgList(thresh), RooArgList(nbkg));
// Do the fit to the sidebands
RooFitResult *fitResult_Sideband = bkgModel.fitTo(data_sideband, Extended(kTRUE), Range("fit"), Verbose(kTRUE), Save(kTRUE), Minos(kFALSE), PrintLevel(3$
bkgModel.plotOn(frame, LineColor(kRed), LineStyle(2), Normalization(nbkg.getVal(), RooAbsReal::NumEvent));
bkgModel.paramOn(frame, ShowConstants(kTRUE), Label("Fit Parameters"), Format("NEU", AutoPrecision(1)));
int status = fitResult_Sideband->status();
int covQual = fitResult_Sideband->covQual();
cout << "The sideband fit status was: " << status << " with a covariance quality code of: " << covQual << endl;
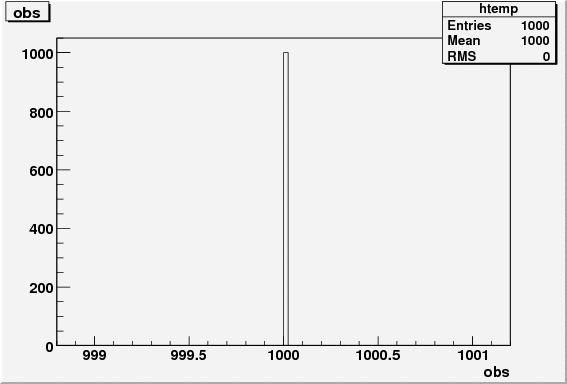
res.obs = sideband_region_count;
res.obs_err = sqrt(res.obs);
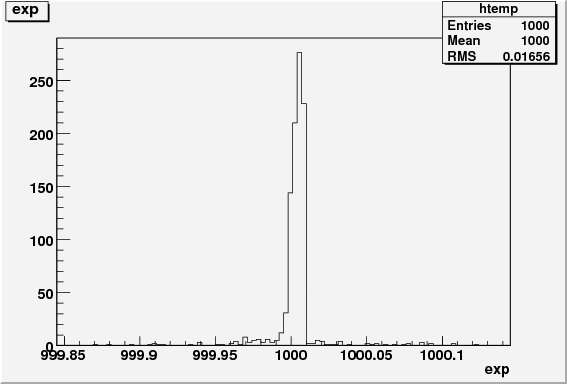
res.exp = nbkg.getVal();
res.exp_err = nbkg.getError();
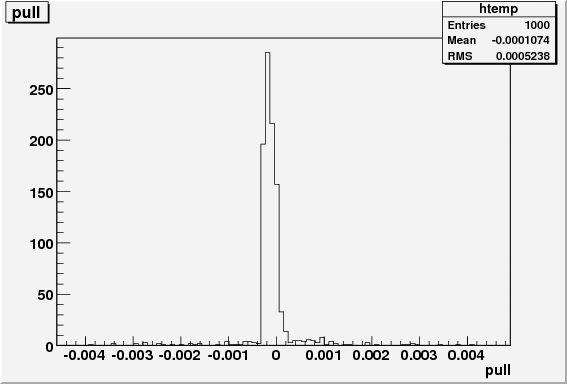
res.delta = res.obs - res.exp;
res.pull = res.delta / res.exp_err;
res.side_fit_status = fitResult_Sideband->status();
res.side_cov_status = fitResult_Sideband->covQual();
res.mean = gauss_mean.getVal();
res.mean_err = gauss_mean.getError();
res.width = gauss_width.getVal();
res.width_err = gauss_width.getError();
return(res);
}I do the above 1000 times and the Results are then saved into a root file for viewing. Now, I would expect to get a nice pull distribution centered at 0 with a width of 1. I do not. I have attached the pull distribution to this post so that you can see it. I have also included as an attachment the full code with all of its spaghetti glory. Is this a by product of me using the extended likelihood technique? Thanks for any help on this.
Justace

code.tar.gz (4.95 KB)