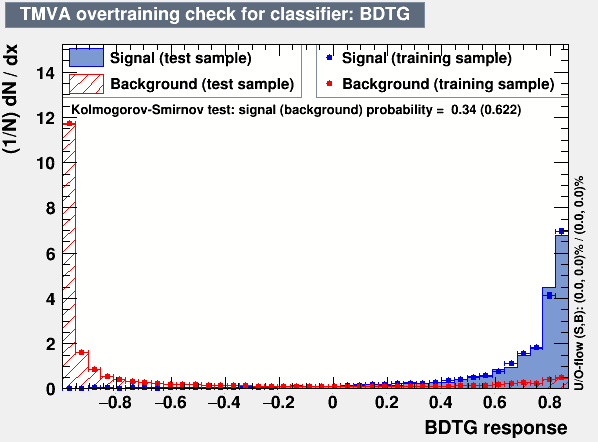
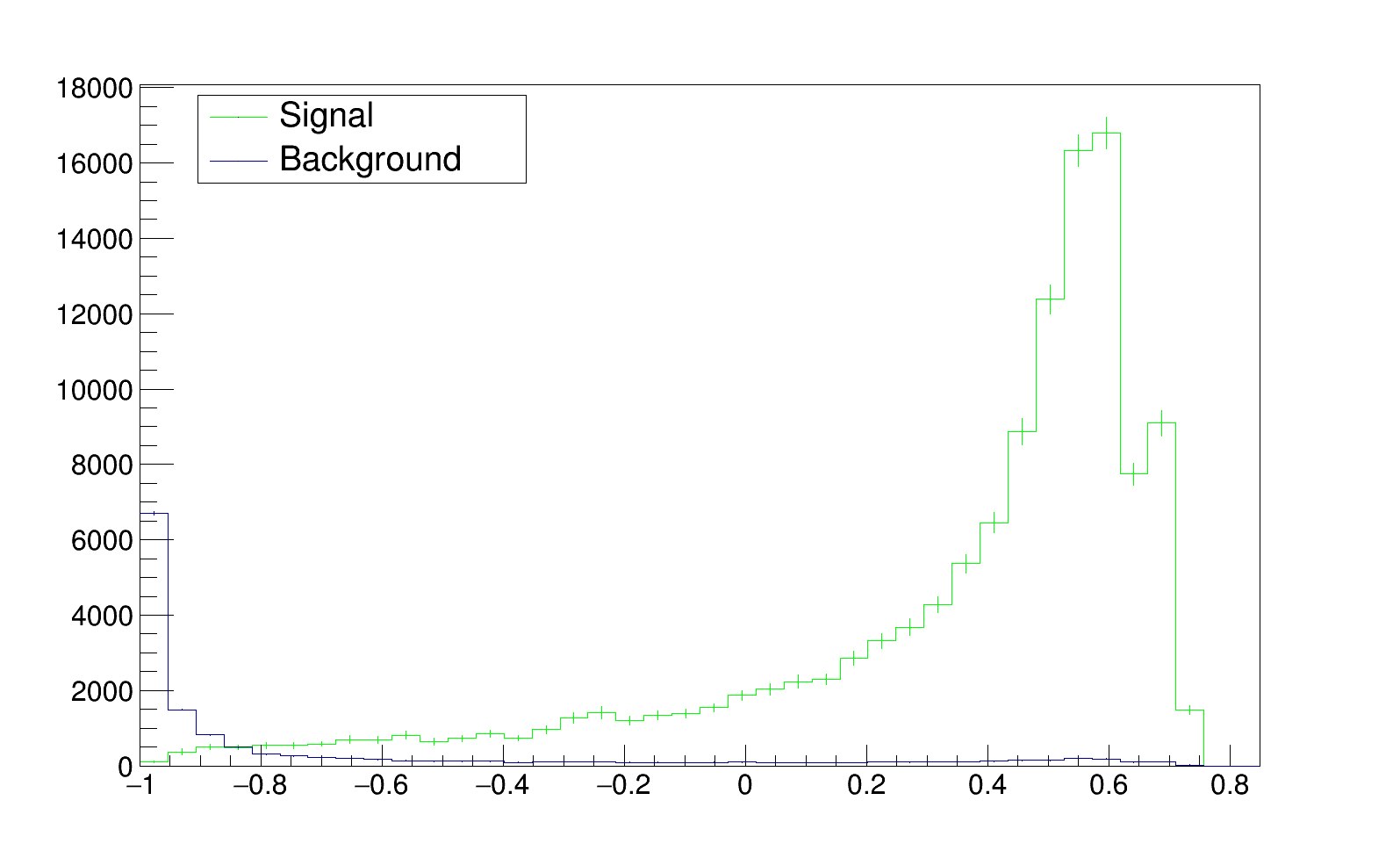
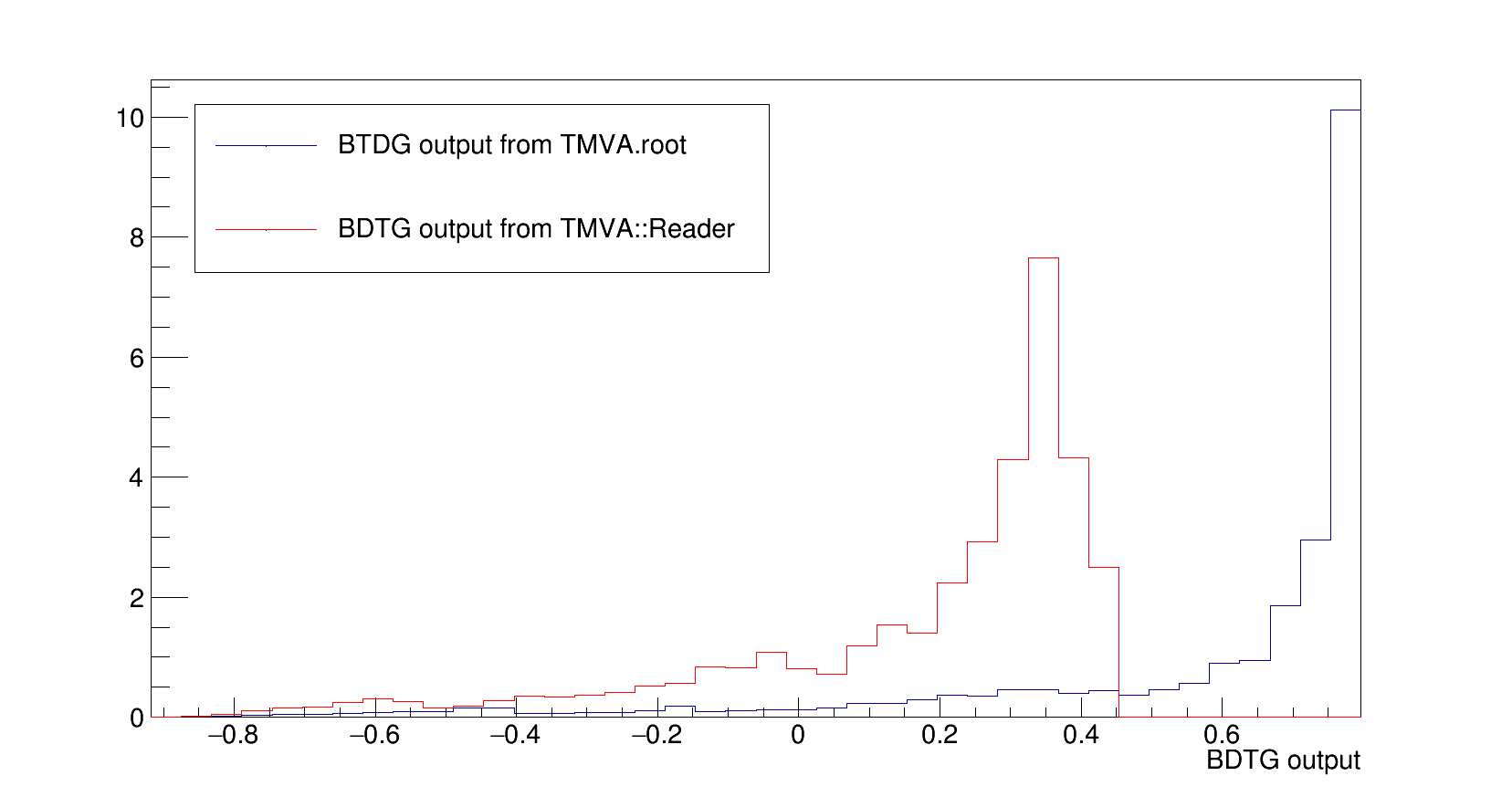
Hi,
For test and training , events are picked randomly from the analysis dataset.
The background is estimated on data and signal on MC. In the analysis, I estimate the shape of the BDTG output in pt bins of the event’s leading photon. for example :
is the BDTG output for one of my pt bins. To produce it I follow the recipe :
Float_t NHiso_photon,Photoniso_photon,E1_3_photon,E2_2_photon,E2_5_photon,E5_5_photon,R9_photon,hadTowOverEm,etawidth_photon,phiwidth_photon,sigmaietaieta_photon;
TMVA::Reader *Access_weight = new TMVA::Reader( “!Color:!Silent” );
Access_weight-> AddVariable( “R9_photon” ,&R9_photon);
Access_weight-> AddVariable( “hadTowOverEm” ,&hadTowOverEm);
Access_weight-> AddVariable( “etawidth_photon” ,&etawidth_photon);
Access_weight-> AddVariable( “phiwidth_photon” ,&phiwidth_photon);
Access_weight-> AddVariable( “sigmaietaieta_photon” ,&sigmaietaieta_photon);
Access_weight-> AddVariable( “NHiso_photon” ,&NHiso_photon);
Access_weight-> AddVariable( “E2_5_photon/E5_5_photon” ,&E2_5_photon);
Access_weight-> AddVariable( “E2_2_photon/E5_5_photon” ,&E2_2_photon);
Access_weight-> AddVariable( “E1_3_photon/E5_5_photon” ,&E1_3_photon);
Access_weight-> AddVariable( “Photoniso_photon” ,&Photoniso_photon);
Access_weight-> BookMVA( “BDTG”, “/dataset/weights/TMVAClassification_BDTG.weights.xml” );
And in the event loop :
NHiso_photon = (Float_t) sig_var_NHiso_photon;
Photoniso_photon = (Float_t)sig_var_Photoniso_photon;
E1_3_photon = (Float_t)sig_var_E1_3_photon/sig_var_E5_5_photon;
E2_2_photon = (Float_t)sig_var_E2_2_photon/sig_var_E5_5_photon;
E2_5_photon = (Float_t)sig_var_E2_5_photon/sig_var_E5_5_photon;
R9_photon = (Float_t)sig_var_R9_photon;
hadTowOverEm = (Float_t)sig_var_hadTowOverEm ;
etawidth_photon = (Float_t)sig_var_etawidth_photon;
phiwidth_photon = (Float_t)sig_var_phiwidth_photon;
sigmaietaieta_photon = (Float_t)sig_var_sigmaietaieta_photon;
Double_t mvaValue = Access_weight->EvaluateMVA( “BDTG” );
I do that for signal and then background. All variables used are ID variable widely used to identify photons across the CMS collaboration.
Concerning the size of the sample in each bin, even in the highly populated bin the distribution are shifted and show a strange behaviour, the bin shown above is one of them.
Let me know if you need more information,
Thanks for your help,
Cheers,
Hugues